Genomic evidence for homoploid hybrid speciation in a marine mammal apex predator
- PMID: 37134171
- PMCID: PMC10156116
- DOI: 10.1126/sciadv.adf6601
Genomic evidence for homoploid hybrid speciation in a marine mammal apex predator
Abstract
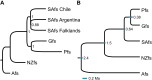
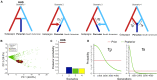
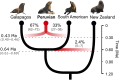
Hybridization is widespread and constitutes an important source of genetic variability and evolution. In animals, its role in generating novel and independent lineages (hybrid speciation) has been strongly debated, with only a few cases supported by genomic data. The South American fur seal (SAfs) Arctocephalus australis is a marine apex predator of Pacific and Atlantic waters, with a disjunct set of populations in Peru and Northern Chile [Peruvian fur seal (Pfs)] with controversial taxonomic status. We demonstrate, using complete genome and reduced representation sequencing, that the Pfs is a genetically distinct species with an admixed genome that originated from hybridization between the SAfs and the Galapagos fur seal (Arctocephalus galapagoensis) ~400,000 years ago. Our results strongly support the origin of Pfs by homoploid hybrid speciation over alternative introgression scenarios. This study highlights the role of hybridization in promoting species-level biodiversity in large vertebrates.
Figures



References
-
- J. Mallet, Hybrid speciation. Nature 446, 279–283 (2007). - PubMed
-
- J. Mallet, Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (2005). - PubMed
-
- R. Abbott, D. Albach, S. Ansell, J. W. Arntzen, S. J. E. Baird, N. Bierne, J. Boughman, A. Brelsford, C. A. Buerkle, R. Buggs, R. K. Butlin, U. Dieckmann, F. Eroukhmanoff, A. Grill, S. H. Cahan, J. S. Hermansen, G. Hewitt, A. G. Hudson, C. Jiggins, J. Jones, B. Keller, T. Marczewski, J. Mallet, P. Martinez-Rodriguez, M. Möst, S. Mullen, R. Nichols, A. W. Nolte, C. Parisod, K. Pfennig, A. M. Rice, M. G. Ritchie, B. Seifert, C. M. Smadja, R. Stelkens, J. M. Szymura, R. Väinölä, J. B. W. Wolf, D. Zinner, Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013). - PubMed
-
- A. Runemark, C. N. Trier, F. Eroukhmanoff, J. S. Hermansen, M. Matschiner, T. O. Elgvin, G. P. Sætre, Variation and constraints in hybrid genome formation. Nat. Ecol. Evol. 2, 549–556 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

