Small Molecules Targeting DNA Polymerase Theta (POLθ) as Promising Synthetic Lethal Agents for Precision Cancer Therapy
- PMID: 37134182
- PMCID: PMC10226047
- DOI: 10.1021/acs.jmedchem.2c02101
Small Molecules Targeting DNA Polymerase Theta (POLθ) as Promising Synthetic Lethal Agents for Precision Cancer Therapy
Abstract
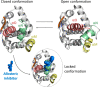
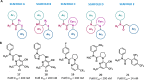
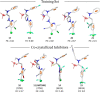
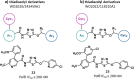
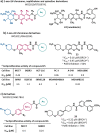
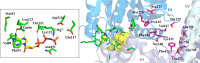
Synthetic lethality (SL) is an innovative strategy in targeted anticancer therapy that exploits tumor genetic vulnerabilities. This topic has come to the forefront in recent years, as witnessed by the increased number of publications since 2007. The first proof of concept for the effectiveness of SL was provided by the approval of poly(ADP-ribose)polymerase inhibitors, which exploit a SL interaction in BRCA-deficient cells, although their use is limited by resistance. Searching for additional SL interactions involving BRCA mutations, the DNA polymerase theta (POLθ) emerged as an exciting target. This review summarizes, for the first time, the POLθ polymerase and helicase inhibitors reported to date. Compounds are described focusing on chemical structure and biological activity. With the aim to enable further drug discovery efforts in interrogating POLθ as a target, we propose a plausible pharmacophore model for POLθ-pol inhibitors and provide a structural analysis of the known POLθ ligand binding sites.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


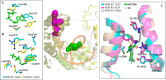
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical
Molecular Biology Databases

