A blueprint for a synthetic genetic feedback optimizer
- PMID: 37137895
- PMCID: PMC10156725
- DOI: 10.1038/s41467-023-37903-0
A blueprint for a synthetic genetic feedback optimizer
Abstract
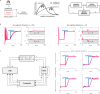
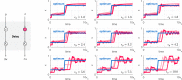
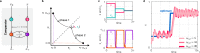
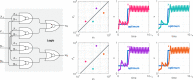
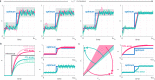
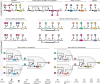
Biomolecular control enables leveraging cells as biomanufacturing factories. Despite recent advancements, we currently lack genetically encoded modules that can be deployed to dynamically fine-tune and optimize cellular performance. Here, we address this shortcoming by presenting the blueprint of a genetic feedback module to optimize a broadly defined performance metric by adjusting the production and decay rate of a (set of) regulator species. We demonstrate that the optimizer can be implemented by combining available synthetic biology parts and components, and that it can be readily integrated with existing pathways and genetically encoded biosensors to ensure its successful deployment in a variety of settings. We further illustrate that the optimizer successfully locates and tracks the optimum in diverse contexts when relying on mass action kinetics-based dynamics and parameter values typical in Escherichia coli.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

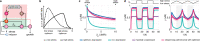
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

