N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry
- PMID: 37137912
- PMCID: PMC10156733
- DOI: 10.1038/s41467-023-38218-w
N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry
Abstract
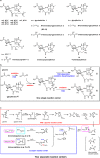
Oxidized cysteine residues are highly reactive and can form functional covalent conjugates, of which the allosteric redox switch formed by the lysine-cysteine NOS bridge is an example. Here, we report a noncanonical FAD-dependent enzyme Orf1 that adds a glycine-derived N-formimidoyl group to glycinothricin to form the antibiotic BD-12. X-ray crystallography was used to investigate this complex enzymatic process, which showed Orf1 has two substrate-binding sites that sit 13.5 Å apart unlike canonical FAD-dependent oxidoreductases. One site could accommodate glycine and the other glycinothricin or glycylthricin. Moreover, an intermediate-enzyme adduct with a NOS-covalent linkage was observed in the later site, where it acts as a two-scissile-bond linkage facilitating nucleophilic addition and cofactor-free decarboxylation. The chain length of nucleophilic acceptors vies with bond cleavage sites at either N-O or O-S accounting for N-formimidoylation or N-iminoacetylation. The resultant product is no longer sensitive to aminoglycoside-modifying enzymes, a strategy that antibiotic-producing species employ to counter drug resistance in competing species.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

