Lung endothelial cells regulate pulmonary fibrosis through FOXF1/R-Ras signaling
- PMID: 37137915
- PMCID: PMC10156846
- DOI: 10.1038/s41467-023-38177-2
Lung endothelial cells regulate pulmonary fibrosis through FOXF1/R-Ras signaling
Abstract
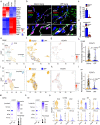
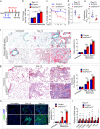
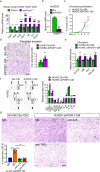
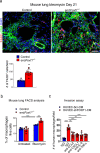
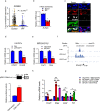
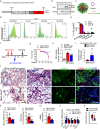
Pulmonary fibrosis results from dysregulated lung repair and involves multiple cell types. The role of endothelial cells (EC) in lung fibrosis is poorly understood. Using single cell RNA-sequencing we identified endothelial transcription factors involved in lung fibrogenesis, including FOXF1, SMAD6, ETV6 and LEF1. Focusing on FOXF1, we found that FOXF1 is decreased in EC within human idiopathic pulmonary fibrosis (IPF) and mouse bleomycin-injured lungs. Endothelial-specific Foxf1 inhibition in mice increased collagen depositions, promoted lung inflammation, and impaired R-Ras signaling. In vitro, FOXF1-deficient EC increased proliferation, invasion and activation of human lung fibroblasts, and stimulated macrophage migration by secreting IL-6, TNFα, CCL2 and CXCL1. FOXF1 inhibited TNFα and CCL2 through direct transcriptional activation of Rras gene promoter. Transgenic overexpression or endothelial-specific nanoparticle delivery of Foxf1 cDNA decreased pulmonary fibrosis in bleomycin-injured mice. Nanoparticle delivery of FOXF1 cDNA can be considered for future therapies in IPF.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Raghu G, et al. Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am. J. Respir. Crit. Care Med. 2018;198:e44–e68. - PubMed
-
- Nogee LM, et al. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N. Engl. J. Med. 2001;344:573–579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

