High efficiency preparation of monodisperse plasma membrane derived extracellular vesicles for therapeutic applications
- PMID: 37137966
- PMCID: PMC10156699
- DOI: 10.1038/s42003-023-04859-2
High efficiency preparation of monodisperse plasma membrane derived extracellular vesicles for therapeutic applications
Abstract
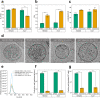
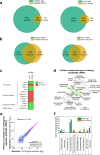
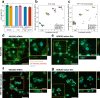
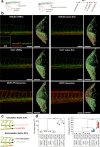
Extracellular vesicles (EVs) are highly interesting for the design of next-generation therapeutics. However, their preparation methods face challenges in standardization, yield, and reproducibility. Here, we describe a highly efficient and reproducible EV preparation method for monodisperse nano plasma membrane vesicles (nPMVs), which yields 10 to 100 times more particles per cell and hour than conventional EV preparation methods. nPMVs are produced by homogenizing giant plasma membrane vesicles following cell membrane blebbing and apoptotic body secretion induced by chemical stressors. nPMVs showed no significant differences compared to native EVs from the same cell line in cryo-TEM analysis, in vitro cellular interactions, and in vivo biodistribution studies in zebrafish larvae. Proteomics and lipidomics, on the other hand, suggested substantial differences consistent with the divergent origin of these two EV types and indicated that nPMVs primarily derive from apoptotic extracellular vesicles. nPMVs may provide an attractive source for developing EV-based pharmaceutical therapeutics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Cell-Derived Vesicles for Antibiotic Delivery-Understanding the Challenges of a Biogenic Carrier System.Small. 2023 Jun;19(25):e2207479. doi: 10.1002/smll.202207479. Epub 2023 Mar 20. Small. 2023. PMID: 36938700
-
Biogenesis, Membrane Trafficking, Functions, and Next Generation Nanotherapeutics Medicine of Extracellular Vesicles.Int J Nanomedicine. 2021 May 18;16:3357-3383. doi: 10.2147/IJN.S310357. eCollection 2021. Int J Nanomedicine. 2021. PMID: 34040369 Free PMC article. Review.
-
Extracellular vesicles from human umbilical cord blood plasma modulate interleukin-2 signaling of T cells to ameliorate experimental autoimmune encephalomyelitis.Theranostics. 2020 Apr 6;10(11):5011-5028. doi: 10.7150/thno.42742. eCollection 2020. Theranostics. 2020. PMID: 32308765 Free PMC article.
-
SIV Infection Regulates Compartmentalization of Circulating Blood Plasma miRNAs within Extracellular Vesicles (EVs) and Extracellular Condensates (ECs) and Decreases EV-Associated miRNA-128.Viruses. 2023 Feb 24;15(3):622. doi: 10.3390/v15030622. Viruses. 2023. PMID: 36992331 Free PMC article.
-
Zebrafish as a preclinical model for Extracellular Vesicle-based therapeutic development.Adv Drug Deliv Rev. 2021 Sep;176:113815. doi: 10.1016/j.addr.2021.05.025. Epub 2021 May 29. Adv Drug Deliv Rev. 2021. PMID: 34058284 Review.
Cited by
-
Stoichiometric constraints for detection of EV-borne biomarkers in blood.J Extracell Vesicles. 2025 Feb;14(2):e70034. doi: 10.1002/jev2.70034. J Extracell Vesicles. 2025. PMID: 39901737 Free PMC article. Review.
-
Extracellular Vesicles: Investigating the Pathophysiology of Diabetes-Associated Hypertension and Diabetic Nephropathy.Biology (Basel). 2023 Aug 16;12(8):1138. doi: 10.3390/biology12081138. Biology (Basel). 2023. PMID: 37627022 Free PMC article. Review.
-
Emerging technologies towards extracellular vesicles large-scale production.Bioact Mater. 2025 Jun 13;52:338-365. doi: 10.1016/j.bioactmat.2025.06.005. eCollection 2025 Oct. Bioact Mater. 2025. PMID: 40585384 Free PMC article. Review.
-
Extracellular Vesicle Mobility in Collagen I Hydrogels Is Influenced by Matrix-Binding Integrins.ACS Nano. 2024 Oct 29;18(43):29585-29601. doi: 10.1021/acsnano.4c07186. Epub 2024 Oct 14. ACS Nano. 2024. PMID: 39400273 Free PMC article.
-
Nano Plasma Membrane Vesicle-Lipid Nanoparticle Hybrids for Enhanced Gene Delivery and Expression.Adv Healthc Mater. 2025 Jan;14(1):e2401888. doi: 10.1002/adhm.202401888. Epub 2024 Nov 10. Adv Healthc Mater. 2025. PMID: 39523736 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

