Systematic identification of endogenous strong constitutive promoters from the diazotrophic rhizosphere bacterium Pseudomonas stutzeri DSM4166 to improve its nitrogenase activity
- PMID: 37138314
- PMCID: PMC10155442
- DOI: 10.1186/s12934-023-02085-3
Systematic identification of endogenous strong constitutive promoters from the diazotrophic rhizosphere bacterium Pseudomonas stutzeri DSM4166 to improve its nitrogenase activity
Abstract
Background: Biological nitrogen fixation converting atmospheric dinitrogen to ammonia is an important way to provide nitrogen for plants. Pseudomonas stutzeri DSM4166 is a diazotrophic Gram-negative bacterium isolated from the rhizosphere of cereal Sorghum nutans. Endogenous constitutive promoters are important for engineering of the nitrogen fixation pathway, however, they have not been systematically characterized in DSM4166.
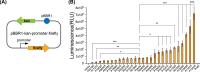
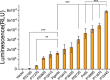
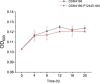
Results: Twenty-six candidate promoters were identified from DSM4166 by RNA-seq analysis. These 26 promoters were cloned and characterized using the firefly luciferase gene. The strengths of nineteen promoters varied from 100 to 959% of the strength of the gentamicin resistance gene promoter. The strongest P12445 promoter was used to overexpress the biological nitrogen fixation pathway-specific positive regulator gene nifA. The transcription level of nitrogen fixation genes in DSM4166 were significantly increased and the nitrogenase activity was enhanced by 4.1 folds determined by the acetylene reduction method. The nifA overexpressed strain produced 359.1 µM of extracellular ammonium which was 25.6 times higher than that produced by the wild-type strain.
Conclusions: The endogenous strong constitutive promoters identified in this study will facilitate development of DSM4166 as a microbial cell factory for nitrogen fixation and production of other useful compounds.
Keywords: Biological nitrogen fixation; Luciferase assay; Metabolic engineering; Promoters; Pseudomonas stutzeri; RNA-seq.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Development of a bacterial gene transcription activating strategy based on transcriptional activator positive feedback.J Adv Res. 2024 Dec;66:155-164. doi: 10.1016/j.jare.2023.12.015. Epub 2023 Dec 18. J Adv Res. 2024. PMID: 38123018 Free PMC article.
-
Complete genome sequence of the nitrogen-fixing and rhizosphere-associated bacterium Pseudomonas stutzeri strain DSM4166.J Bacteriol. 2011 Jul;193(13):3422-3. doi: 10.1128/JB.05039-11. Epub 2011 Apr 22. J Bacteriol. 2011. PMID: 21515765 Free PMC article.
-
The novel regulatory ncRNA, NfiS, optimizes nitrogen fixation via base pairing with the nitrogenase gene nifK mRNA in Pseudomonas stutzeri A1501.Proc Natl Acad Sci U S A. 2016 Jul 26;113(30):E4348-56. doi: 10.1073/pnas.1604514113. Epub 2016 Jul 12. Proc Natl Acad Sci U S A. 2016. PMID: 27407147 Free PMC article.
-
Molecular Mechanism and Agricultural Application of the NifA-NifL System for Nitrogen Fixation.Int J Mol Sci. 2023 Jan 4;24(2):907. doi: 10.3390/ijms24020907. Int J Mol Sci. 2023. PMID: 36674420 Free PMC article. Review.
-
Azotobacters as biofertilizer.Adv Appl Microbiol. 2019;108:1-43. doi: 10.1016/bs.aambs.2019.07.001. Epub 2019 Aug 28. Adv Appl Microbiol. 2019. PMID: 31495403 Review.
Cited by
-
Pangenomic landscapes shape performances of a synthetic genetic circuit across Stutzerimonas species.mSystems. 2024 Sep 17;9(9):e0084924. doi: 10.1128/msystems.00849-24. Epub 2024 Aug 21. mSystems. 2024. PMID: 39166875 Free PMC article.
-
Development of a bacterial gene transcription activating strategy based on transcriptional activator positive feedback.J Adv Res. 2024 Dec;66:155-164. doi: 10.1016/j.jare.2023.12.015. Epub 2023 Dec 18. J Adv Res. 2024. PMID: 38123018 Free PMC article.
-
Enhanced extracellular ammonium release in the plant endophyte Gluconacetobacter diazotrophicus through genome editing.Microbiol Spectr. 2024 Jan 11;12(1):e0247823. doi: 10.1128/spectrum.02478-23. Epub 2023 Dec 1. Microbiol Spectr. 2024. PMID: 38038458 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

