A westernized diet changed the colonic bacterial composition and metabolite concentration in a dextran sulfate sodium pig model for ulcerative colitis
- PMID: 37138607
- PMCID: PMC10150118
- DOI: 10.3389/fmicb.2023.1018242
A westernized diet changed the colonic bacterial composition and metabolite concentration in a dextran sulfate sodium pig model for ulcerative colitis
Erratum in
-
Corrigendum: A westernized diet changed the colonic bacterial composition and metabolite concentration in a dextran sulfate sodium pig model for ulcerative colitis.Front Microbiol. 2023 Jun 27;14:1238157. doi: 10.3389/fmicb.2023.1238157. eCollection 2023. Front Microbiol. 2023. PMID: 37440879 Free PMC article.
Abstract
Introduction: Ulcerative colitis (UC) is characterized by chronic inflammation in the colonic epithelium and has a blurred etiology. A western diet and microbial dysbiosis in the colon were reported to play a role in UC development. In this study, we investigated the effect of a westernized diet, i.e., increasing fat and protein content by including ground beef, on the colonic bacterial composition in a dextran sulfate sodium (DexSS) challenged pig study.
Methods: The experiment was carried out in three complete blocks following a 2×2 factorial design including 24 six-week old pigs, fed either a standard diet (CT) or the standard diet substituted with 15% ground beef to simulate a typical westernized diet (WD). Colitis was induced in half of the pigs on each dietary treatment by oral administration of DexSS (DSS and WD+DSS, respectively). Samples from proximal and distal colon and feces were collected.
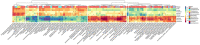
Results and discussion: Bacterial alpha diversity was unaffected by experimental block, and sample type. In proximal colon, WD group had similar alpha diversity to CT group and the WD+DSS group showed the lowest alpha diversity compared to the other treatment groups. There was a significant interaction between western diet and DexSS for beta diversity, based on Bray-Curtis dissimilarly. The westernized diet and DexSS resulted in three and seven differentially abundant phyla, 21 and 65 species, respectively, mainly associated with the Firmicutes and Bacteroidota phyla followed by Spirochaetota, Desulfobacterota, and Proteobacteria. The concentration of short-chain fatty acids (SCFA) was lowest in the distal colon. Treatment had a slight effect on the estimates for microbial metabolites that might have valuable biological relevance for future studies. The concentration of putrescine in the colon and feces and that of total biogenic amines was highest in the WD+DSS group. We conclude that a westernized diet could be a potential risk factor and an exacerbating agent for UC by reducing the abundance of SCFA-producing bacteria, increasing the abundance of pathogens such as Helicobacter trogontum, and by increasing the concentration of microbial proteolytic-derived metabolites in the colon.
Keywords: 16S rRNA gut metagenomics; colonic inflammation; dextran sulfate sodium; inflammatory bowel disease; meat consumption; porcine model; ulcerative colitis.
Copyright © 2023 Panah, Nielsen, Simpson, Schönherz, Schramm, Lauridsen, Nielsen, Højberg, Fredborg, Purup and Canibe.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- AOAC (1990). Official methods of analysis. AOAC, Washington, DC.
-
- Banaszkiewicz A., Kądzielska J., Gawrońska A., Pituch H., Obuch-Woszczatyński P., Albrecht P., et al. . (2014). Enterotoxigenic Clostridium perfringens infection and pediatric patients with inflammatory bowel disease. J. Crohn's Colitis 8, 276–281. doi: 10.1016/j.crohns.2013.08.018, PMID: - DOI - PubMed
-
- Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01 - DOI
LinkOut - more resources
Full Text Sources

