Human milk-associated bacterial communities associate with the infant gut microbiome over the first year of life
- PMID: 37138613
- PMCID: PMC10149717
- DOI: 10.3389/fmicb.2023.1164553
Human milk-associated bacterial communities associate with the infant gut microbiome over the first year of life
Abstract
Introduction: Microbial communities inhabiting the human infant gut are important for immune system development and lifelong health. One critical exposure affecting the bacterial colonization of the infant gut is consumption of human milk, which contains diverse microbial communities and prebiotics. We hypothesized that human milk-associated microbial profiles are associated with those of the infant gut.
Methods: Maternal-infant dyads enrolled in the New Hampshire Birth Cohort Study (n = 189 dyads) contributed breast milk and infant stool samples collected approximately at 6 weeks, 4 months, 6 months, 9 months, and 12 months postpartum (n = 572 samples). Microbial DNA was extracted from milk and stool and the V4-V5 region of the bacterial 16S rRNA gene was sequenced.
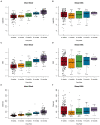
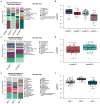
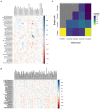
Results: Clustering analysis identified three breast milk microbiome types (BMTs), characterized by differences in Streptococcus, Staphylococcus, Pseudomonas, Acinetobacter, and microbial diversity. Four 6-week infant gut microbiome types (6wIGMTs) were identified, differing in abundances of Bifidobacterium, Bacteroides, Clostridium, Streptococcus, and Escherichia/Shigella, while two 12-month IGMTs (12mIGMTs) differed primarily by Bacteroides presence. At 6 weeks, BMT was associated with 6wIGMT (Fisher's exact test value of p = 0.039); this association was strongest among infants delivered by Cesarean section (Fisher's exact test value of p = 0.0028). The strongest correlations between overall breast milk and infant stool microbial community structures were observed when comparing breast milk samples to infant stool samples collected at a subsequent time point, e.g., the 6-week breast milk microbiome associated with the 6-month infant gut microbiome (Mantel test Z-statistic = 0.53, value of p = 0.001). Streptoccous and Veillonella species abundance were correlated in 6-week milk and infant stool, and 4- and 6-month milk Pantoea species were associated with infant stool Lachnospiraceae genera at 9 and 12 months.
Discussion: We identified clusters of human milk and infant stool microbial communities that were associated in maternal-infant dyads at 6 weeks of life and found that milk microbial communities were more strongly associated with infant gut microbial communities in infants delivered operatively and after a lag period. These results suggest that milk microbial communities have a long-term effect on the infant gut microbiome both through sharing of microbes and other molecular mechanisms.
Keywords: breast milk microbiome; breastfeeding; cesarean delivery; infant gut microbiome; microbial co-occurrence.
Copyright © 2023 Lundgren, Madan, Karagas, Morrison, Christensen and Hoen.
Conflict of interest statement
SL conducted this research while at the Geisel School of Medicine at Dartmouth and is currently affiliated with Nightingale Health Plc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner.Microbiome. 2018 Jul 5;6(1):109. doi: 10.1186/s40168-018-0490-8. Microbiome. 2018. PMID: 29973274 Free PMC article.
-
Breastmilk, Stool, and Meconium: Bacterial Communities in South Africa.Microb Ecol. 2022 Jan;83(1):246-251. doi: 10.1007/s00248-021-01758-z. Epub 2021 Apr 22. Microb Ecol. 2022. PMID: 33885917 Free PMC article.
-
Evolution of the Gut Microbiome in HIV-Exposed Uninfected and Unexposed Infants during the First Year of Life.mBio. 2022 Oct 26;13(5):e0122922. doi: 10.1128/mbio.01229-22. Epub 2022 Sep 8. mBio. 2022. PMID: 36073815 Free PMC article.
-
Comparing early life nutritional sources and human milk feeding practices: personalized and dynamic nutrition supports infant gut microbiome development and immune system maturation.Gut Microbes. 2023 Jan-Dec;15(1):2190305. doi: 10.1080/19490976.2023.2190305. Gut Microbes. 2023. PMID: 37055920 Free PMC article. Review.
-
A narrative review of the functional components of human breast milk and their potential to modulate the gut microbiome, the consideration of maternal and child characteristics, and confounders of breastfeeding, and their impact on risk of obesity later in life.Nutr Rev. 2023 Apr 11;81(5):597-609. doi: 10.1093/nutrit/nuac072. Nutr Rev. 2023. PMID: 36048515 Review.
Cited by
-
Feeding Expressed Breast Milk Alters the Microbial Network of Breast Milk and Increases Breast Milk Microbiome Diversity over Time.Microorganisms. 2024 Dec 25;13(1):12. doi: 10.3390/microorganisms13010012. Microorganisms. 2024. PMID: 39858780 Free PMC article.
-
Early-life and concurrent predictors of the healthy adolescent microbiome in a cohort study.Genome Med. 2025 May 8;17(1):50. doi: 10.1186/s13073-025-01481-1. Genome Med. 2025. PMID: 40340756 Free PMC article.
-
Infant gut microbiota and SCFAs mediate the association between early-life human milk microbiota and neurodevelopment.NPJ Biofilms Microbiomes. 2025 Aug 1;11(1):149. doi: 10.1038/s41522-025-00790-y. NPJ Biofilms Microbiomes. 2025. PMID: 40750606 Free PMC article.
-
Bacterial Community of Breast Milk in Breastfeeding Women Using Culture-Dependent and Culture-Independent Approaches.J Microbiol Biotechnol. 2024 Oct 28;34(10):2005-2011. doi: 10.4014/jmb.2407.07001. Epub 2024 Sep 9. J Microbiol Biotechnol. 2024. PMID: 39252644 Free PMC article.
-
Role of Human Milk Microbiota in Infant Neurodevelopment: Mechanisms and Clinical Implications.Children (Basel). 2024 Nov 30;11(12):1476. doi: 10.3390/children11121476. Children (Basel). 2024. PMID: 39767905 Free PMC article. Review.
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources

