The power of DNA based methods in probiotic authentication
- PMID: 37138639
- PMCID: PMC10150049
- DOI: 10.3389/fmicb.2023.1158440
The power of DNA based methods in probiotic authentication
Abstract
Introduction: The global probiotic market is growing rapidly, and strict quality control measures are required to ensure probiotic product efficacy and safety. Quality assurance of probiotic products involve confirming the presence of specific probiotic strains, determining the viable cell counts, and confirming the absence of contaminant strains. Third-party evaluation of probiotic quality and label accuracy is recommended for probiotic manufacturers. Following this recommendation, multiple batches of a top selling multi-strain probiotic product were evaluated for label accuracy.
Methods: A total of 55 samples (five multi-strain finished products and 50 single-strain raw ingredients) containing a total of 100 probiotic strains were evaluated using a combination of molecular methods including targeted PCR, non-targeted amplicon-based High Throughput Sequencing (HTS), and non-targeted Shotgun Metagenomic Sequencing (SMS).
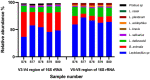
Results: Targeted testing using species-specific or strain-specific PCR methods confirmed the identity of all strains/species. While 40 strains were identified to strain level, 60 strains were identified to species level only due to lack of strain-specific identification methods. In amplicon based HTS, two variable regions of 16S rRNA gene were targeted. Based on V5-V8 region data, ~99% of total reads per sample corresponded to target species, and no undeclared species were detected. Based on V3-V4 region data, ~95%-97% of total reads per sample corresponded to target species, while ~2%-3% of reads matched undeclared species (Proteus species), however, attempts to culture Proteus confirmed that all batches were free from viable Proteus species. Reads from SMS assembled to the genomes of all 10 target strains in all five batches of the finished product.
Discussion: While targeted methods enable quick and accurate identification of target taxa in probiotic products, non-targeted methods enable the identification of all species in a product including undeclared species, with the caveats of complexity, high cost, and long time to result.
Keywords: amplicon-based high throughput sequencing; authentication; probiotic; shotgun metagenomic sequencing; species-specific; strain-specific.
Copyright © 2023 Shehata and Newmaster.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

