Clustering of Cardiac Transcriptome Profiles Reveals Unique: Subgroups of Dilated Cardiomyopathy Patients
- PMID: 37138803
- PMCID: PMC10149655
- DOI: 10.1016/j.jacbts.2022.10.010
Clustering of Cardiac Transcriptome Profiles Reveals Unique: Subgroups of Dilated Cardiomyopathy Patients
Abstract
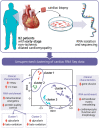
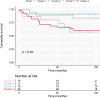
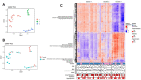
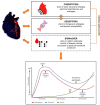
Dilated cardiomyopathy is a heterogeneous disease characterized by multiple genetic and environmental etiologies. The majority of patients are treated the same despite these differences. The cardiac transcriptome provides information on the patient's pathophysiology, which allows targeted therapy. Using clustering techniques on data from the genotype, phenotype, and cardiac transcriptome of patients with early- and end-stage dilated cardiomyopathy, more homogeneous patient subgroups are identified based on shared underlying pathophysiology. Distinct patient subgroups are identified based on differences in protein quality control, cardiac metabolism, cardiomyocyte function, and inflammatory pathways. The identified pathways have the potential to guide future treatment and individualize patient care.
Keywords: clustering; dilated cardiomyopathy; genetics; transcriptomics.
© 2023 The Authors.
Conflict of interest statement
The research leading to these results has received funding from the DCVA DOUBLE DOSE grant. We acknowledge the support from the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation and CVON Arena-PRIME, 2017-18. Drs Verdonschot and Nabben are supported by a Dutch Heart Foundation grant. Heymans receives personal fees for scientific advice to Astra-Zeneca, Pfizer, Novo Nordisk and CSL Behringer. All other authors have reported that they have no relationships relevant to the contents of this paper.
Figures




References
-
- Japp A.G., Gulati A., Cook S.A., Cowie M.R., Prasad S.K. The diagnosis and evaluation of dilated cardiomyopathy. J Am Coll Cardiol. 2016;67:2996–3010. - PubMed
-
- Verdonschot J.A.J., Derks K.W.J., Hazebroek M.R., et al. Distinct cardiac transcriptomic clustering in titin and lamin A/C-associated dilated cardiomyopathy patients. Circulation. 2020;142:1230–1232. - PubMed
-
- Verdonschot J.A.J., Hazebroek M.R., Krapels I.P.C., et al. Implications of genetic testing in dilated cardiomyopathy. Circ Genom Precis Med. 2020;13:476–487. - PubMed
LinkOut - more resources
Full Text Sources

