A Nomogram for Predicting Delayed Viral Shedding in Non-Severe SARS-CoV-2 Omicron Infection
- PMID: 37138833
- PMCID: PMC10150765
- DOI: 10.2147/IDR.S407620
A Nomogram for Predicting Delayed Viral Shedding in Non-Severe SARS-CoV-2 Omicron Infection
Abstract
Purpose: The Omicron variant of SARS-CoV-2 has emerged as a significant global concern, characterized by its rapid transmission and resistance to existing treatments and vaccines. However, the specific hematological and biochemical factors that may impact the clearance of Omicron variant infection remain unclear. The present study aimed to identify easily accessible laboratory markers that are associated with prolonged virus shedding in non-severe patients with COVID-19 caused by the Omicron variant.
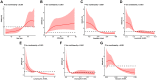
Patients and methods: A retrospective cohort study was conducted on 882 non-severe COVID-19 patients who were diagnosed with the Omicron variant in Shanghai between March and June 2022. The least absolute shrinkage and selection operator regression model was used for feature selection and dimensional reduction, and multivariate logistic regression analysis was performed to construct a nomogram for predicting the risk of prolonged SARS-CoV-2 RNA positivity lasting for more than 7 days. The receiver operating characteristic (ROC) curve and calibration curves were used to assess predictive discrimination and accuracy, with bootstrap validation.
Results: Patients were randomly divided into derivation (70%, n = 618) and validation (30%, n = 264) cohorts. Optimal independent markers for prolonged viral shedding time (VST) over 7 days were identified as Age, C-reactive protein (CRP), platelet count, leukocyte count, lymphocyte count, and eosinophil count. These factors were subsequently incorporated into the nomogram utilizing bootstrap validation. The area under the curve (AUC) in the derivation (0.761) and validation (0.756) cohorts indicated good discriminative ability. The calibration curve showed good agreement between the nomogram-predicted and actual patients with VST over 7 days.
Conclusion: Our study confirmed six factors associated with delayed VST in non-severe SARS-CoV-2 Omicron infection and constructed a Nomogram which may assist non-severely affected patients to better estimate the appropriate length of self-isolation and optimize their self-management strategies.
Keywords: COVID-19; Omicron; SARS-CoV-2; nomogram; viral shedding time.
© 2023 Yu et al.
Conflict of interest statement
The authors no conflicts of interest in this work.
Figures



Similar articles
-
Development and validation of a nomogram to predict failure of 14-day negative nucleic acid conversion in adults with non-severe COVID-19 during the Omicron surge: a retrospective multicenter study.Infect Dis Poverty. 2023 Feb 7;12(1):7. doi: 10.1186/s40249-023-01057-4. Infect Dis Poverty. 2023. PMID: 36750862 Free PMC article.
-
Development and validation of an individualized nomogram for early prediction of the duration of SARS-CoV-2 shedding in COVID-19 patients with non-severe disease.J Zhejiang Univ Sci B. 2021 Apr 15;22(4):318-329. doi: 10.1631/jzus.B2000608. J Zhejiang Univ Sci B. 2021. PMID: 33835766 Free PMC article.
-
Association between immunity and viral shedding duration in non-severe SARS-CoV-2 Omicron variant-infected patients.Front Public Health. 2022 Dec 22;10:1032957. doi: 10.3389/fpubh.2022.1032957. eCollection 2022. Front Public Health. 2022. PMID: 36620263 Free PMC article.
-
Comorbidities prolonged viral shedding of patients infected with SARS-CoV-2 omicron variant in Shanghai: A multi-center, retrospective, observational study.J Infect Public Health. 2023 Feb;16(2):182-189. doi: 10.1016/j.jiph.2022.12.003. Epub 2022 Dec 6. J Infect Public Health. 2023. PMID: 36566602 Free PMC article.
-
Duration of viable virus shedding and polymerase chain reaction positivity of the SARS-CoV-2 Omicron variant in the upper respiratory tract: a systematic review and meta-analysis.Int J Infect Dis. 2023 Apr;129:228-235. doi: 10.1016/j.ijid.2023.02.011. Epub 2023 Feb 18. Int J Infect Dis. 2023. PMID: 36804640 Free PMC article.
Cited by
-
A Nomogram for Predicting Survival in Patients with SARS-CoV-2 Omicron Variant Pneumonia Based on Admission Data.Infect Drug Resist. 2025 Apr 25;18:2093-2104. doi: 10.2147/IDR.S509178. eCollection 2025. Infect Drug Resist. 2025. PMID: 40303607 Free PMC article.
-
SARS-CoV-2 Omicron variant infection affects blood platelets, a comparative analysis with Delta variant.Front Immunol. 2023 Sep 27;14:1231576. doi: 10.3389/fimmu.2023.1231576. eCollection 2023. Front Immunol. 2023. PMID: 37828997 Free PMC article.
-
Associations between genetic mutations in different SARS-CoV-2 strains and negative conversion time of viral RNA among imported cases in Hangzhou: A cross-sectional study.Virus Res. 2024 Jul;345:199400. doi: 10.1016/j.virusres.2024.199400. Epub 2024 May 20. Virus Res. 2024. PMID: 38763300 Free PMC article.
-
Retrospective Analysis of Factors Associated with a Prolonged Nucleic Acid Conversion Time in Patients with COVID-19 at Fangcang Shelter Hospital.Infect Drug Resist. 2025 Jun 30;18:3219-3229. doi: 10.2147/IDR.S521808. eCollection 2025. Infect Drug Resist. 2025. PMID: 40620275 Free PMC article.
-
Predictive value of systemic inflammatory index (SII) for the time to negative nucleic acid conversion in patients with mild COVID-19 by the omicron wave.Front Med (Lausanne). 2025 May 1;11:1474236. doi: 10.3389/fmed.2024.1474236. eCollection 2024. Front Med (Lausanne). 2025. PMID: 40376291 Free PMC article.
References
-
- Prasad N, Derado G, Nanduri SA, et al. Effectiveness of a COVID-19 additional primary or booster vaccine dose in preventing SARS-CoV-2 infection among nursing home residents during widespread circulation of the omicron variant — United States, February 14–March 27, 2022. Morbidity Mortal Wkly Rep. 2022;71(18):633–637. doi:10.15585/mmwr.mm7118a4 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

