The performance of metagenomic next-generation sequencing in diagnosing pulmonary infectious diseases using authentic clinical specimens: The Illumina platform versus the Beijing Genomics Institute platform
- PMID: 37138853
- PMCID: PMC10149716
- DOI: 10.3389/fphar.2023.1164633
The performance of metagenomic next-generation sequencing in diagnosing pulmonary infectious diseases using authentic clinical specimens: The Illumina platform versus the Beijing Genomics Institute platform
Abstract
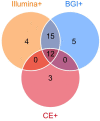
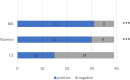
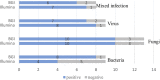
Introduction: Metagenomic next-generation sequencing (mNGS) has been increasingly used to detect infectious organisms and is rapidly moving from research to clinical laboratories. Presently, mNGS platforms mainly include those from Illumina and the Beijing Genomics Institute (BGI). Previous studies have reported that various sequencing platforms have similar sensitivity in detecting the reference panel that mimics clinical specimens. However, whether the Illumina and BGI platforms provide the same diagnostic performance using authentic clinical samples remains unclear. Methods: In this prospective study, we compared the performance of the Illumina and BGI platforms in detecting pulmonary pathogens. Forty-six patients with suspected pulmonary infection were enrolled in the final analysis. All patients received bronchoscopy, and the specimens collected were sent for mNGS on the two different sequencing platforms. Results: The diagnostic sensitivity of the Illumina and BGI platforms was notably higher than that of conventional examination (76.9% vs. 38.5%, p < 0.001; 82.1% vs. 38.5%, p < 0.001; respectively). The sensitivity and specificity for pulmonary infection diagnosis were not significantly different between the Illumina and BGI platforms. Furthermore, the pathogenic detection rate of the two platforms were not significantly different. Conclusion: The Illumina and BGI platforms exhibited similar diagnostic performance for pulmonary infectious diseases using clinical specimens, and both are superior to conventional examinations.
Keywords: BGI; Illumina; conventional examination; metagenomic next-generation sequencing; pulmonary infection.
Copyright © 2023 Han, Zhao, Yang, Huang, Wang and Feng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Diagnostic performance of the metagenomic next-generation sequencing in lung biopsy tissues in patients suspected of having a local pulmonary infection.BMC Pulm Med. 2022 Mar 29;22(1):112. doi: 10.1186/s12890-022-01912-4. BMC Pulm Med. 2022. PMID: 35351079 Free PMC article.
-
Illumina and Nanopore sequencing in culture-negative samples from suspected lower respiratory tract infection patients.Front Cell Infect Microbiol. 2024 Apr 4;14:1230650. doi: 10.3389/fcimb.2024.1230650. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38638824 Free PMC article.
-
[Analysis of the methods and quality assurance of metagenomic next-generation sequencing to detect the microbial cfDNA from blood samples in China].Zhonghua Yi Xue Za Zhi. 2022 Apr 19;102(15):1114-1118. doi: 10.3760/cma.j.cn112137-20220104-00017. Zhonghua Yi Xue Za Zhi. 2022. PMID: 35436811 Chinese.
-
The clinical application of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid: case reports and literature review.Expert Rev Mol Diagn. 2022 May;22(5):575-582. doi: 10.1080/14737159.2022.2071607. Epub 2022 May 2. Expert Rev Mol Diagn. 2022. PMID: 35473493 Review.
-
A Comprehensive Review of Performance of Next-Generation Sequencing Platforms.Biomed Res Int. 2022 Sep 29;2022:3457806. doi: 10.1155/2022/3457806. eCollection 2022. Biomed Res Int. 2022. Retraction in: Biomed Res Int. 2024 Jan 9;2024:9837802. doi: 10.1155/2024/9837802. PMID: 36212714 Free PMC article. Retracted. Review.
Cited by
-
Evaluation of the diagnostic utility of metagenomic next-generation sequencing testing for pathogen identification in infected hosts: a retrospective cohort study.Ther Adv Infect Dis. 2024 Feb 23;11:20499361241232854. doi: 10.1177/20499361241232854. eCollection 2024 Jan-Dec. Ther Adv Infect Dis. 2024. PMID: 38404751 Free PMC article.
References
LinkOut - more resources
Full Text Sources

