Next generation immuno-oncology tumor profiling using a rapid, non-invasive, computational biophysics biomarker in early-stage breast cancer
- PMID: 37138891
- PMCID: PMC10149754
- DOI: 10.3389/frai.2023.1153083
Next generation immuno-oncology tumor profiling using a rapid, non-invasive, computational biophysics biomarker in early-stage breast cancer
Abstract
Background: Immuno-oncology (IO) therapies targeting the PD-1/PD-L1 axis, such as immune checkpoint inhibitor (ICI) antibodies, have emerged as promising treatments for early-stage breast cancer (ESBC). Despite immunotherapy's clinical significance, the number of benefiting patients remains small, and the therapy can prompt severe immune-related events. Current pathologic and transcriptomic predictions of IO response are limited in terms of accuracy and rely on single-site biopsies, which cannot fully account for tumor heterogeneity. In addition, transcriptomic analyses are costly and time-consuming. We therefore constructed a computational biomarker coupling biophysical simulations and artificial intelligence-based tissue segmentation of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRIs), enabling IO response prediction across the entire tumor.
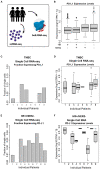
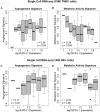
Methods: By analyzing both single-cell and whole-tissue RNA-seq data from non-IO-treated ESBC patients, we associated gene expression levels of the PD-1/PD-L1 axis with local tumor biology. PD-L1 expression was then linked to biophysical features derived from DCE-MRIs to generate spatially- and temporally-resolved atlases (virtual tumors) of tumor biology, as well as the TumorIO biomarker of IO response. We quantified TumorIO within patient virtual tumors (n = 63) using integrative modeling to train and develop a corresponding TumorIO Score.
Results: We validated the TumorIO biomarker and TumorIO Score in a small, independent cohort of IO-treated patients (n = 17) and correctly predicted pathologic complete response (pCR) in 15/17 individuals (88.2% accuracy), comprising 10/12 in triple negative breast cancer (TNBC) and 5/5 in HR+/HER2- tumors. We applied the TumorIO Score in a virtual clinical trial (n = 292) simulating ICI administration in an IO-naïve cohort that underwent standard chemotherapy. Using this approach, we predicted pCR rates of 67.1% for TNBC and 17.9% for HR+/HER2- tumors with addition of IO therapy; comparing favorably to empiric pCR rates derived from published trials utilizing ICI in both cancer subtypes.
Conclusion: The TumorIO biomarker and TumorIO Score represent a next generation approach using integrative biophysical analysis to assess cancer responsiveness to immunotherapy. This computational biomarker performs as well as PD-L1 transcript levels in identifying a patient's likelihood of pCR following anti-PD-1 IO therapy. The TumorIO biomarker allows for rapid IO profiling of tumors and may confer high clinical decision impact to further enable personalized oncologic care.
Keywords: ESBC; ICI; biophysical simulation; computational biomarker; immune checkpoint inhibitor; immuno-oncology; virtual clinical trial; virtual tumors.
Copyright © 2023 Cook, Biancalana, Liadis, Lopez Ramos, Zhang, Patel, Peterson, Pfeiffer, Cole and Antony.
Conflict of interest statement
All authors are employed by SimBioSys, Inc. and are involved in the commercial development of the Simul-omics 4D Engine, the TumorIO biomarker, and the TumorIO Score.
Figures





References
-
- Bonsang-Kitzis H., Sadacca B., Hamy-Petit A. S., Moarii M., Pinheiro A., Laurent C., et al. . (2016). Biological network-driven gene selection identifies a stromal immune module as a key determinant of triple-negative breast carcinoma prognosis. Oncoimmunology 5, 1061176. 10.1080/2162402X.2015.1061176 - DOI - PMC - PubMed
-
- Caballo M., Sanderink W. B. G., Han L., Gao Y., Athanasiou A., Mann R. M., et al. . (2022). Four-dimensional machine learning radiomics for the pretreatment assessment of breast cancer pathologic complete response to neoadjuvant chemotherapy in dynamic contrast-enhanced MRI. J. Magnet. Reson. Imag. 2022, 28273. 10.1002/jmri.28273 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

