The regulatory mechanisms and inhibitors of isocitrate dehydrogenase 1 in cancer
- PMID: 37139412
- PMCID: PMC10149907
- DOI: 10.1016/j.apsb.2022.12.019
The regulatory mechanisms and inhibitors of isocitrate dehydrogenase 1 in cancer
Abstract
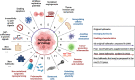
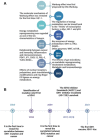
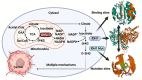
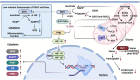
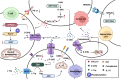
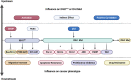
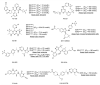
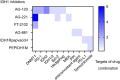
Reprogramming of energy metabolism is one of the basic characteristics of cancer and has been proved to be an important cancer treatment strategy. Isocitrate dehydrogenases (IDHs) are a class of key proteins in energy metabolism, including IDH1, IDH2, and IDH3, which are involved in the oxidative decarboxylation of isocitrate to yield α-ketoglutarate (α-KG). Mutants of IDH1 or IDH2 can produce d-2-hydroxyglutarate (D-2HG) with α-KG as the substrate, and then mediate the occurrence and development of cancer. At present, no IDH3 mutation has been reported. The results of pan-cancer research showed that IDH1 has a higher mutation frequency and involves more cancer types than IDH2, implying IDH1 as a promising anti-cancer target. Therefore, in this review, we summarized the regulatory mechanisms of IDH1 on cancer from four aspects: metabolic reprogramming, epigenetics, immune microenvironment, and phenotypic changes, which will provide guidance for the understanding of IDH1 and exploring leading-edge targeted treatment strategies. In addition, we also reviewed available IDH1 inhibitors so far. The detailed clinical trial results and diverse structures of preclinical candidates illustrated here will provide a deep insight into the research for the treatment of IDH1-related cancers.
Keywords: Cancer; D-2HG; Epigenetics; IDH1; IDH1 inhibitors; Immune microenvironment; Metabolic reprogramming; Regulatory mechanisms.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Conflict of interest statement
All the authors declared that they have no conflicts of interest.
Figures



References
-
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. - PubMed
-
- DeBerardinis R.J., Chandel N.S. We need to talk about the Warburg effect. Nat Metab. 2020;2:127–129. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

