A genome-wide association study of morphometric traits in dromedaries
- PMID: 37139670
- PMCID: PMC10357240
- DOI: 10.1002/vms3.1151
A genome-wide association study of morphometric traits in dromedaries
Abstract
Background: Investigating genomic regions associated with morphometric traits in camels is valuable, because it allows a better understanding of adaptive and productive features to implement a sustainable management and a customised breeding program for dromedaries.
Objectives: With a genome-wide association study (GWAS) including 96 Iranian dromedaries phenotyped for 12 morphometric traits and genotyped-by-sequencing (GBS) with 14,522 SNPs, we aimed at identifying associated candidate genes.
Methods: The association between SNPs and morphometric traits was investigated using a linear mixed model with principal component analysis (PCA) and kinship matrix.
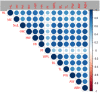
Results: With this approach, we detected 59 SNPs located in 37 candidate genes potentially associated to morphometric traits in dromedaries. The top associated SNPs were related to pin width, whither to pin length, height at whither, muzzle girth, and tail length. Interestingly, the results highlight the association between whither height, muzzle circumference, tail length, whither to pin length. The identified candidate genes were associated with growth, body size, and immune system in other species.
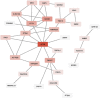
Conclusions: We identified three key hub genes in the gene network analysis including ACTB, SOCS1 and ARFGEF1. In the central position of gene network, ACTB was detected as the most important gene related to muscle function. With this initial GWAS using GBS on dromedary camels for morphometric traits, we show that this SNP panel can be effective for genetic evaluation of growth in dromedaries. However, we suggest a higher-density SNP array may greatly improve the reliability of the results.
Keywords: Camelus dromedarius; candidate genes; genotyping by sequencing; single nucleotide polymorphism (SNP).
© 2023 The Authors. Veterinary Medicine and Science published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Identification of novel genetic loci related to dromedary camel (Camelus dromedarius) morphometrics, biomechanics, and behavior by genome-wide association studies.BMC Vet Res. 2024 Sep 18;20(1):418. doi: 10.1186/s12917-024-04263-w. BMC Vet Res. 2024. PMID: 39294626 Free PMC article.
-
Genomic prediction for growth using a low-density SNP panel in dromedary camels.Sci Rep. 2021 Apr 7;11(1):7675. doi: 10.1038/s41598-021-87296-7. Sci Rep. 2021. PMID: 33828208 Free PMC article.
-
A multi-breed GWAS for morphometric traits in four Beninese indigenous cattle breeds reveals loci associated with conformation, carcass and adaptive traits.BMC Genomics. 2020 Nov 11;21(1):783. doi: 10.1186/s12864-020-07170-0. BMC Genomics. 2020. PMID: 33176675 Free PMC article.
-
Genome-Wide Diversity, Population Structure and Demographic History of Dromedaries in the Central Desert of Iran.Genes (Basel). 2020 May 29;11(6):599. doi: 10.3390/genes11060599. Genes (Basel). 2020. PMID: 32485848 Free PMC article.
-
Development and validation of the Axiom-MaruPri SNP chip for genetic analyses of domesticated old world camelids.Gene. 2024 Aug 30;921:148541. doi: 10.1016/j.gene.2024.148541. Epub 2024 May 8. Gene. 2024. PMID: 38723784
Cited by
-
The related SNPs and genes to body size using GWAS- latent variable modeling in dromedaries.BMC Genomics. 2025 Jul 8;26(1):645. doi: 10.1186/s12864-025-11766-9. BMC Genomics. 2025. PMID: 40629317 Free PMC article.
-
Genomic patterns of selection in morphometric traits across diverse Indian cattle breeds.Mamm Genome. 2024 Sep;35(3):377-389. doi: 10.1007/s00335-024-10047-2. Epub 2024 Jul 16. Mamm Genome. 2024. PMID: 39014170
-
Identification of novel genetic loci related to dromedary camel (Camelus dromedarius) morphometrics, biomechanics, and behavior by genome-wide association studies.BMC Vet Res. 2024 Sep 18;20(1):418. doi: 10.1186/s12917-024-04263-w. BMC Vet Res. 2024. PMID: 39294626 Free PMC article.
References
-
- Abdelmanova, A. S. , Sermyagin, A. A. , Dotsev, A. V. , Bardukov, N. V. , Fornara, M. S. , Brem, G. , & Zinovieva, N. A. (2022). Genome‐wide screening for SNPs associated with stature in diverse cattle breeds. Diversity, 14, 692.
-
- Al‐Mamun, H. A. , Kwan, P. , Clark, S. A. , Ferdosi, M. H. , Tellam, R. , & Gondro, C. (2015). Genome‐wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genetics, Selection, Evolution., 47, 1–11. - PMC - PubMed
-
- An, B. , Xia, J. , Chang, T. , Wang, X. , Xu, L. , Zhang, L. , Gao, X. , Chen, Y. , Li, J. , & Gao, H. (2019). Genome‐wide association study reveals candidate genes associated with body measurement traits in Chinese Wagyu beef cattle. Animal Genetics, 50, 386–390. - PubMed
-
- Barazandeh, A. , Mohammadabadi, M. R. , Ghaderi, M. , & Nezamabadipour, H. (2016). Genome‐wide analysis of CpG islands in some livestock genomes and their relationship with genomic features. Czech Journal of Animal Science, 61, e487.
-
- Barazandeh, A. , Mohammadabadi, M. R. , Ghaderi‐Zefrehei, M. , & Rafeied, F. (2019). Whole genome comparative analysis of CpG islands in camelid and other mammalian genomes. Mammalian Biology, 98, 73–79.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

