A Kiss of Deep Homology: Partial Convergence in the Genomic Basis of Hypertrophied Lips in Cichlid Fish and Human Cleft Lip
- PMID: 37140021
- PMCID: PMC10195091
- DOI: 10.1093/gbe/evad072
A Kiss of Deep Homology: Partial Convergence in the Genomic Basis of Hypertrophied Lips in Cichlid Fish and Human Cleft Lip
Abstract
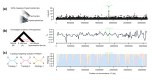
The genomic loci generating both adaptive and maladaptive variation could be surprisingly predictable in deeply homologous vertebrate structures like the lips. Variation in highly conserved vertebrate traits such as the jaws and teeth in organisms as evolutionarily disparate as teleost fishes and mammals is known to be structured by the same genes. Likewise, hypertrophied lips that have evolved repeatedly in Neotropical and African cichlid fish lineages could share unexpectedly similar genetic bases themselves and even provide surprising insight into the loci underlying human craniofacial anomalies. To isolate the genomic regions underlying adaptive divergence in hypertrophied lips, we first employed genome-wide associations (GWAs) in several species of African cichlids from Lake Malawi. Then, we tested if these GWA regions were shared through hybridization with another Lake Malawi cichlid lineage that has evolved hypertrophied lips seemingly in parallel. Overall, introgression among hypertrophied lip lineages appeared limited. Among our Malawi GWA regions, one contained the gene kcnj2 that has been implicated in the convergently evolved hypertrophied lips in Central American Midas cichlids that diverged from the Malawi radiation over 50 million years ago. The Malawi hypertrophied lip GWA regions also contained several additional genes that cause human lip-associated birth defects. Cichlid fishes are becoming prominent examples of replicated genomic architecture underlying trait convergence and are increasingly providing insight into human craniofacial anomalies such as a cleft lip.
Keywords: animal model; evolutionary medicine; genome resequencing; human birth defects.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Ansari M, et al. . 2014. A syndromic form of Pierre Robin sequence is caused by 5q23 deletions encompassing FBN2 and PHAX. Eur J Med Genet. 57(10):587–595. - PubMed
-
- Arnegard ME, Snoeks J. 2001. New three-spotted cichlid species with hypertrophied lips (Teleostei: Cichlidae) from the deep waters of Lake Malaŵi/Nyasa, Africa. Copeia 2001:705–717.
-
- Baumgarten L, Machado-Schiaffino G, Henning F, Meyer A. 2015. What big lips are good for: on the adaptive function of repeatedly evolved hypertrophied lips of cichlid fishes. Biol J Linn Soc. 115:448–455.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

