Proksee: in-depth characterization and visualization of bacterial genomes
- PMID: 37140037
- PMCID: PMC10320063
- DOI: 10.1093/nar/gkad326
Proksee: in-depth characterization and visualization of bacterial genomes
Abstract
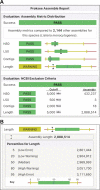
Proksee (https://proksee.ca) provides users with a powerful, easy-to-use, and feature-rich system for assembling, annotating, analysing, and visualizing bacterial genomes. Proksee accepts Illumina sequence reads as compressed FASTQ files or pre-assembled contigs in raw, FASTA, or GenBank format. Alternatively, users can supply a GenBank accession or a previously generated Proksee map in JSON format. Proksee then performs assembly (for raw sequence data), generates a graphical map, and provides an interface for customizing the map and launching further analysis jobs. Notable features of Proksee include unique and informative assembly metrics provided via a custom reference database of assemblies; a deeply integrated high-performance genome browser for viewing and comparing analysis results at individual base resolution (developed specifically for Proksee); an ever-growing list of embedded analysis tools whose results can be seamlessly added to the map or searched and explored in other formats; and the option to export graphical maps, analysis results, and log files for data sharing and research reproducibility. All these features are provided via a carefully designed multi-server cloud-based system that can easily scale to meet user demand and that ensures the web server is robust and responsive.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D.et al. .. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012; 19:455–477. - PMC - PubMed
-
- Vallenet D., Calteau A., Dubois M., Amours P., Bazin A., Beuvin M., Burlot L., Bussell X., Fouteau S., Gautreau G.et al. .. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020; 48:D579–D589. - PMC - PubMed
-
- Olson R.D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J.J., Dempsey D.M., Dickerman A., Dietrich E.M., Kenyon R.W.et al. .. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023; 51:D678–D689. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

