Basement membrane proteins in extracellular matrix characterize NF1 neurofibroma development and response to MEK inhibitor
- PMID: 37140985
- PMCID: PMC10266775
- DOI: 10.1172/JCI168227
Basement membrane proteins in extracellular matrix characterize NF1 neurofibroma development and response to MEK inhibitor
Abstract
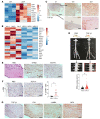
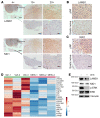
Neurofibromatosis type 1 (NF1) is one of the most common tumor-predisposing genetic disorders. Neurofibromas are NF1-associated benign tumors. A hallmark feature of neurofibromas is an abundant collagen-rich extracellular matrix (ECM) that constitutes more than 50% of the tumor dry weight. However, little is known about the mechanism underlying ECM deposition during neurofibroma development and treatment response. We performed a systematic investigation of ECM enrichment during plexiform neurofibroma (pNF) development and identified basement membrane (BM) proteins, rather than major collagen isoforms, as the most upregulated ECM component. Following MEK inhibitor treatment, the ECM profile displayed an overall downregulation signature, suggesting ECM reduction as a therapeutic benefit of MEK inhibition. Through these proteomic studies, TGF-β1 signaling was identified as playing a role in ECM dynamics. Indeed, TGF-β1 overexpression promoted pNF progression in vivo. Furthermore, by integrating single-cell RNA sequencing, we found that immune cells including macrophages and T cells produce TGF-β1 to induce Schwann cells to produce and deposit BM proteins for ECM remodeling. Following Nf1 loss, neoplastic Schwann cells further increased BM protein deposition in response to TGF-β1. Our data delineate the regulation governing ECM dynamics in pNF and suggest that BM proteins could serve as biomarkers for disease diagnosis and treatment response.
Keywords: Extracellular matrix; Macrophages; Oncology; Tumor suppressors.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

