Recruitment of the SNX17-Retriever recycling pathway regulates synaptic function and plasticity
- PMID: 37141105
- PMCID: PMC10165670
- DOI: 10.1083/jcb.202207025
Recruitment of the SNX17-Retriever recycling pathway regulates synaptic function and plasticity
Abstract
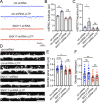
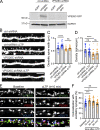
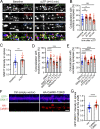
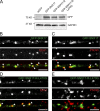
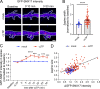
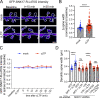
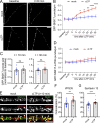
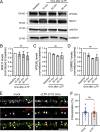
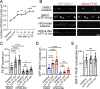
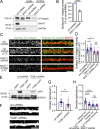
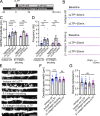
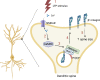
Trafficking of cell-surface proteins from endosomes to the plasma membrane is a key mechanism to regulate synaptic function. In non-neuronal cells, proteins recycle to the plasma membrane either via the SNX27-Retromer-WASH pathway or via the recently discovered SNX17-Retriever-CCC-WASH pathway. While SNX27 is responsible for the recycling of key neuronal receptors, the roles of SNX17 in neurons are less understood. Here, using cultured hippocampal neurons, we demonstrate that the SNX17 pathway regulates synaptic function and plasticity. Disruption of this pathway results in a loss of excitatory synapses and prevents structural plasticity during chemical long-term potentiation (cLTP). cLTP drives SNX17 recruitment to synapses, where its roles are in part mediated by regulating the surface expression of β1-integrin. SNX17 recruitment relies on NMDAR activation, CaMKII signaling, and requires binding to the Retriever and PI(3)P. Together, these findings provide molecular insights into the regulation of SNX17 at synapses and define key roles for SNX17 in synaptic maintenance and in regulating enduring forms of synaptic plasticity.
© 2023 Rivero-Ríos et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



References
-
- Babayan, A.H., Kramár E.A., Barrett R.M., Jafari M., Häettig J., Chen L.Y., Rex C.S., Lauterborn J.C., Wood M.A., Gall C.M., and Lynch G.. 2012. Integrin dynamics produce a delayed stage of long-term potentiation and memory consolidation. J. Neurosci. 32:12854–12861. 10.1523/JNEUROSCI.2024-12.2012 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

