An interferon gamma response signature links myocardial aging and immunosenescence
- PMID: 37141306
- PMCID: PMC10651211
- DOI: 10.1093/cvr/cvad068
An interferon gamma response signature links myocardial aging and immunosenescence
Abstract
Aims: Aging entails profound immunological transformations that can impact myocardial homeostasis and predispose to heart failure. However, preclinical research in the immune-cardiology field is mostly conducted in young healthy animals, which potentially weakens its translational relevance. Herein, we sought to investigate how the aging T-cell compartment associates with changes in myocardial cell biology in aged mice.
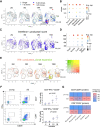
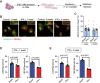
Methods and results: We phenotyped the antigen-experienced effector/memory T cells purified from heart-draining lymph nodes of 2-, 6-, 12-, and 18-month-old C57BL/6J mice using single-cell RNA/T cell receptor sequencing. Simultaneously, we profiled all non-cardiomyocyte cell subsets purified from 2- to 18-month-old hearts and integrated our data with publicly available cardiomyocyte single-cell sequencing datasets. Some of these findings were confirmed at the protein level by flow cytometry. With aging, the heart-draining lymph node and myocardial T cells underwent clonal expansion and exhibited an up-regulated pro-inflammatory transcription signature, marked by an increased interferon-γ (IFN-γ) production. In parallel, all major myocardial cell populations showed increased IFN-γ responsive signature with aging. In the aged cardiomyocytes, a stronger IFN-γ response signature was paralleled by the dampening of expression levels of transcripts related to most metabolic pathways, especially oxidative phosphorylation. Likewise, induced pluripotent stem cells-derived cardiomyocytes exposed to chronic, low grade IFN-γ treatment showed a similar inhibition of metabolic activity.
Conclusions: By investigating the paired age-related alterations in the T cells found in the heart and its draining lymph nodes, we provide evidence for increased myocardial IFN-γ signaling with age, which is associated with inflammatory and metabolic shifts typically seen in heart failure.
Keywords: Cardiac aging; Heart failure and immune cells; Interferon gamma; T cells.
© The Author(s) 2023. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: None declared.
Figures



Comment in
-
Interferon-gamma signs off an old heart.Cardiovasc Res. 2023 Nov 15;119(14):2387-2389. doi: 10.1093/cvr/cvad151. Cardiovasc Res. 2023. PMID: 37883721 No abstract available.
References
-
- Swirski FK, Nahrendorf M. Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat Rev Immunol 2018;18:733–744. - PubMed
-
- Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci 2000;908:244–254. - PubMed
-
- Bollini S, Guzik TJ. Old, but gold? Not the case for the immune system when promoting systemic ageing. Cardiovasc Res 2022;118:e14–e16. - PubMed
-
- Min H, Montecino-Rodriguez E, Dorshkind K. Reduction in the developmental potential of intrathymic T cell progenitors with age. J Immunol 2004;173:245–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

