Intratumor heterogeneity and cell secretome promote chemotherapy resistance and progression of colorectal cancer
- PMID: 37142595
- PMCID: PMC10160076
- DOI: 10.1038/s41419-023-05806-z
Intratumor heterogeneity and cell secretome promote chemotherapy resistance and progression of colorectal cancer
Abstract
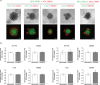
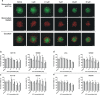
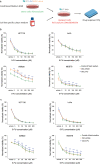
The major underlying cause for the high mortality rate in colorectal cancer (CRC) relies on its drug resistance, to which intratumor heterogeneity (ITH) contributes substantially. CRC tumors have been reported to comprise heterogeneous populations of cancer cells that can be grouped into 4 consensus molecular subtypes (CMS). However, the impact of inter-cellular interaction between these cellular states on the emergence of drug resistance and CRC progression remains elusive. Here, we explored the interaction between cell lines belonging to the CMS1 (HCT116 and LoVo) and the CMS4 (SW620 and MDST8) in a 3D coculture model, mimicking the ITH of CRC. The spatial distribution of each cell population showed that CMS1 cells had a preference to grow in the center of cocultured spheroids, while CMS4 cells localized at the periphery, in line with observations in tumors from CRC patients. Cocultures of CMS1 and CMS4 cells did not alter cell growth, but significantly sustained the survival of both CMS1 and CMS4 cells in response to the front-line chemotherapeutic agent 5-fluorouracil (5-FU). Mechanistically, the secretome of CMS1 cells exhibited a remarkable protective effect for CMS4 cells against 5-FU treatment, while promoting cellular invasion. Secreted metabolites may be responsible for these effects, as demonstrated by the existence of 5-FU induced metabolomic shifts, as well as by the experimental transfer of the metabolome between CMS1 and CMS4 cells. Overall, our results suggest that the interplay between CMS1 and CMS4 cells stimulates CRC progression and reduces the efficacy of chemotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–75. doi: 10.1016/S0140-6736(17)33326-3. - DOI - PMC - PubMed
-
- Colorectal Cancer Survival Rates | Colorectal Cancer Prognosis. https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-st....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

