SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool
- PMID: 37142791
- PMCID: PMC10185471
- DOI: 10.1038/s41556-023-01138-4
SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool
Abstract
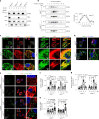
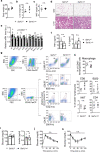
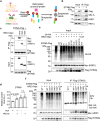
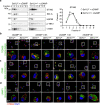
Stimulator of interferon genes (STING) orchestrates the production of proinflammatory cytokines in response to cytosolic double-stranded DNA; however, the pathophysiological significance and molecular mechanism underlying the folding and maturation of nascent STING protein at the endoplasmic reticulum (ER) remain unknown. Here we report that the SEL1L-HRD1 protein complex-the most conserved branch of ER-associated degradation (ERAD)-is a negative regulator of the STING innate immunity by ubiquitinating and targeting nascent STING protein for proteasomal degradation in the basal state. SEL1L or HRD1 deficiency in macrophages specifically amplifies STING signalling and immunity against viral infection and tumour growth. Mechanistically, nascent STING protein is a bona fide substrate of SEL1L-HRD1 in the basal state, uncoupled from ER stress or its sensor inositol-requiring enzyme 1α. Hence, our study not only establishes a key role of SEL1L-HRD1 ERAD in innate immunity by limiting the size of the activable STING pool, but identifies a regulatory mechanism and therapeutic approach to targeting STING.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












Comment in
-
ERADication of STING limits inflammation.Nat Cell Biol. 2023 May;25(5):635-636. doi: 10.1038/s41556-023-01142-8. Nat Cell Biol. 2023. PMID: 37142792 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

