Gut-specific telomerase expression counteracts systemic aging in telomerase-deficient zebrafish
- PMID: 37142828
- PMCID: PMC10191862
- DOI: 10.1038/s43587-023-00401-5
Gut-specific telomerase expression counteracts systemic aging in telomerase-deficient zebrafish
Abstract
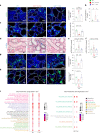
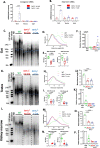
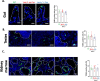
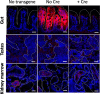
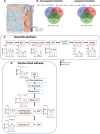
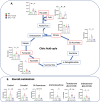
Telomere shortening is a hallmark of aging and is counteracted by telomerase. As in humans, the zebrafish gut is one of the organs with the fastest rate of telomere decline, triggering early tissue dysfunction during normal zebrafish aging and in prematurely aged telomerase mutants. However, whether telomere-dependent aging of an individual organ, the gut, causes systemic aging is unknown. Here we show that tissue-specific telomerase expression in the gut can prevent telomere shortening and rescues premature aging of tert-/-. Induction of telomerase rescues gut senescence and low cell proliferation, while restoring tissue integrity, inflammation and age-dependent microbiota dysbiosis. Averting gut aging causes systemic beneficial impacts, rescuing aging of distant organs such as reproductive and hematopoietic systems. Conclusively, we show that gut-specific telomerase expression extends the lifespan of tert-/- by 40%, while ameliorating natural aging. Our work demonstrates that gut-specific rescue of telomerase expression leading to telomere elongation is sufficient to systemically counteract aging in zebrafish.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

