Parallel sequencing of extrachromosomal circular DNAs and transcriptomes in single cancer cells
- PMID: 37142849
- PMCID: PMC10181933
- DOI: 10.1038/s41588-023-01386-y
Parallel sequencing of extrachromosomal circular DNAs and transcriptomes in single cancer cells
Abstract
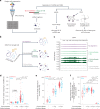
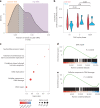
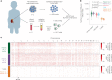
Extrachromosomal DNAs (ecDNAs) are common in cancer, but many questions about their origin, structural dynamics and impact on intratumor heterogeneity are still unresolved. Here we describe single-cell extrachromosomal circular DNA and transcriptome sequencing (scEC&T-seq), a method for parallel sequencing of circular DNAs and full-length mRNA from single cells. By applying scEC&T-seq to cancer cells, we describe intercellular differences in ecDNA content while investigating their structural heterogeneity and transcriptional impact. Oncogene-containing ecDNAs were clonally present in cancer cells and drove intercellular oncogene expression differences. In contrast, other small circular DNAs were exclusive to individual cells, indicating differences in their selection and propagation. Intercellular differences in ecDNA structure pointed to circular recombination as a mechanism of ecDNA evolution. These results demonstrate scEC&T-seq as an approach to systematically characterize both small and large circular DNA in cancer cells, which will facilitate the analysis of these DNA elements in cancer and beyond.
© 2023. The Author(s).
Conflict of interest statement
R.P.K. and A.G.H. are founders of Econic Biosciences Ltd.
Figures




References
-
- Stuart T, Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

