Targeting homologous recombination deficiency in uterine leiomyosarcoma
- PMID: 37143137
- PMCID: PMC10157936
- DOI: 10.1186/s13046-023-02687-0
Targeting homologous recombination deficiency in uterine leiomyosarcoma
Abstract
Background: Uterine leiomyosarcoma (uLMS) is a rare and aggressive gynaecological malignancy, with individuals with advanced uLMS having a five-year survival of < 10%. Mutations in the homologous recombination (HR) DNA repair pathway have been observed in ~ 10% of uLMS cases, with reports of some individuals benefiting from poly (ADP-ribose) polymerase (PARP) inhibitor (PARPi) therapy, which targets this DNA repair defect. In this report, we screened individuals with uLMS, accrued nationally, for mutations in the HR repair pathway and explored new approaches to therapeutic targeting.
Methods: A cohort of 58 individuals with uLMS were screened for HR Deficiency (HRD) using whole genome sequencing (WGS), whole exome sequencing (WES) or NGS panel testing. Individuals identified to have HRD uLMS were offered PARPi therapy and clinical outcome details collected. Patient-derived xenografts (PDX) were generated for therapeutic targeting.
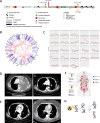
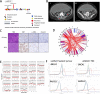
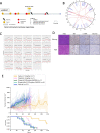
Results: All 13 uLMS samples analysed by WGS had a dominant COSMIC mutational signature 3; 11 of these had high genome-wide loss of heterozygosity (LOH) (> 0.2) but only two samples had a CHORD score > 50%, one of which had a homozygous pathogenic alteration in an HR gene (deletion in BRCA2). A further three samples harboured homozygous HRD alterations (all deletions in BRCA2), detected by WES or panel sequencing, with 5/58 (9%) individuals having HRD uLMS. All five individuals gained access to PARPi therapy. Two of three individuals with mature clinical follow up achieved a complete response or durable partial response (PR) with the subsequent addition of platinum to PARPi upon minor progression during initial PR on PARPi. Corresponding PDX responses were most rapid, complete and sustained with the PARP1-specific PARPi, AZD5305, compared with either olaparib alone or olaparib plus cisplatin, even in a paired sample of a BRCA2-deleted PDX, derived following PARPi therapy in the patient, which had developed PARPi-resistance mutations in PRKDC, encoding DNA-PKcs.
Conclusions: Our work demonstrates the value of identifying HRD for therapeutic targeting by PARPi and platinum in individuals with the aggressive rare malignancy, uLMS and suggests that individuals with HRD uLMS should be included in trials of PARP1-specific PARPi.
Keywords: Homologous recombination deficiency; PARP inhibitors; Patient-derived xenografts; Rare cancers; Uterine leiomyosarcoma.
© 2023. The Author(s).
Conflict of interest statement
CLS declares Advisory Boards for AstraZeneca, Clovis Oncology, Roche, Eisai Inc, Sierra Oncology, Takeda, MSD and Grant/Research support from AstraZeneca, Clovis Oncology, Eisai Inc, Sierra Oncology, Boehringer Ingelheim, Roche and Beigene. Other authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

