Choroid plexus mis-splicing and altered cerebrospinal fluid composition in myotonic dystrophy type 1
- PMID: 37143315
- PMCID: PMC10545633
- DOI: 10.1093/brain/awad148
Choroid plexus mis-splicing and altered cerebrospinal fluid composition in myotonic dystrophy type 1
Abstract
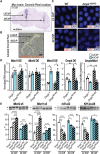
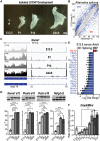
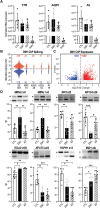
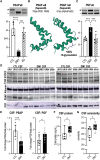
Myotonic dystrophy type 1 is a dominantly inherited multisystemic disease caused by CTG tandem repeat expansions in the DMPK 3' untranslated region. These expanded repeats are transcribed and produce toxic CUG RNAs that sequester and inhibit activities of the MBNL family of developmental RNA processing factors. Although myotonic dystrophy is classified as a muscular dystrophy, the brain is also severely affected by an unusual cohort of symptoms, including hypersomnia, executive dysfunction, as well as early onsets of tau/MAPT pathology and cerebral atrophy. To address the molecular and cellular events that lead to these pathological outcomes, we recently generated a mouse Dmpk CTG expansion knock-in model and identified choroid plexus epithelial cells as particularly affected by the expression of toxic CUG expansion RNAs. To determine if toxic CUG RNAs perturb choroid plexus functions, alternative splicing analysis was performed on lateral and hindbrain choroid plexi from Dmpk CTG knock-in mice. Choroid plexus transcriptome-wide changes were evaluated in Mbnl2 knockout mice, a developmental-onset model of myotonic dystrophy brain dysfunction. To determine if transcriptome changes also occurred in the human disease, we obtained post-mortem choroid plexus for RNA-seq from neurologically unaffected (two females, three males; ages 50-70 years) and myotonic dystrophy type 1 (one female, three males; ages 50-70 years) donors. To test that choroid plexus transcriptome alterations resulted in altered CSF composition, we obtained CSF via lumbar puncture from patients with myotonic dystrophy type 1 (five females, five males; ages 35-55 years) and non-myotonic dystrophy patients (three females, four males; ages 26-51 years), and western blot and osmolarity analyses were used to test CSF alterations predicted by choroid plexus transcriptome analysis. We determined that CUG RNA induced toxicity was more robust in the lateral choroid plexus of Dmpk CTG knock-in mice due to comparatively higher Dmpk and lower Mbnl RNA levels. Impaired transitions to adult splicing patterns during choroid plexus development were identified in Mbnl2 knockout mice, including mis-splicing previously found in Dmpk CTG knock-in mice. Whole transcriptome analysis of myotonic dystrophy type 1 choroid plexus revealed disease-associated RNA expression and mis-splicing events. Based on these RNA changes, predicted alterations in ion homeostasis, secretory output and CSF composition were confirmed by analysis of myotonic dystrophy type 1 CSF. Our results implicate choroid plexus spliceopathy and concomitant alterations in CSF homeostasis as an unappreciated contributor to myotonic dystrophy type 1 CNS pathogenesis.
Keywords: RNA disease; alternative splicing; development; neurodegeneration; short tandem repeat expansion.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
M.S.S. is a Scientific Advisory Board member for Kate Therapeutics, Skyhawk Therapeutics and Tacit Therapeutics.
Figures


References
-
- Weskamp K, Olwin BB, Parker R. Post-transcriptional regulation in skeletal muscle development, repair, and disease. Trends Mol Med. 2021;27:469–481. - PubMed
-
- Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

