Metabarcoding of soil environmental DNA to estimate plant diversity globally
- PMID: 37143888
- PMCID: PMC10151745
- DOI: 10.3389/fpls.2023.1106617
Metabarcoding of soil environmental DNA to estimate plant diversity globally
Abstract
Introduction: Traditional approaches to collecting large-scale biodiversity data pose huge logistical and technical challenges. We aimed to assess how a comparatively simple method based on sequencing environmental DNA (eDNA) characterises global variation in plant diversity and community composition compared with data derived from traditional plant inventory methods.
Methods: We sequenced a short fragment (P6 loop) of the chloroplast trnL intron from from 325 globally distributed soil samples and compared estimates of diversity and composition with those derived from traditional sources based on empirical (GBIF) or extrapolated plant distribution and diversity data.
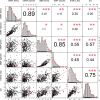
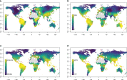
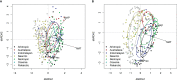
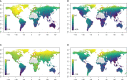
Results: Large-scale plant diversity and community composition patterns revealed by sequencing eDNA were broadly in accordance with those derived from traditional sources. The success of the eDNA taxonomy assignment, and the overlap of taxon lists between eDNA and GBIF, was greatest at moderate to high latitudes of the northern hemisphere. On average, around half (mean: 51.5% SD 17.6) of local GBIF records were represented in eDNA databases at the species level, depending on the geographic region.
Discussion: eDNA trnL gene sequencing data accurately represent global patterns in plant diversity and composition and thus can provide a basis for large-scale vegetation studies. Important experimental considerations for plant eDNA studies include using a sampling volume and design to maximise the number of taxa detected and optimising the sequencing depth. However, increasing the coverage of reference sequence databases would yield the most significant improvements in the accuracy of taxonomic assignments made using the P6 loop of the trnL region.
Keywords: TRNL; distribution; diversity; environmental DNA; molecular methods; plant; soil.
Copyright © 2023 Vasar, Davison, Moora, Sepp, Anslan, Al-Quraishy, Bahram, Bueno, Cantero, Fabiano, Decocq, Drenkhan, Fraser, Oja, Garibay-Orijel, Hiiesalu, Koorem, Mucina, Öpik, Põlme, Pärtel, Phosri, Semchenko, Vahter, Doležal, Palacios, Tedersoo and Zobel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Barnes C. J., Nielsen I. B., Aagaard A., Ejrnæs R., Hansen A. J., Frøslev T. G. (2022). Metabarcoding of soil environmental DNA replicates plant community variation but not specificity. Environ. DNA 4 (4), 732–746. doi: 10.1002/edn3.287 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

