Pericyte-derived cells participate in optic nerve scar formation
- PMID: 37143930
- PMCID: PMC10151493
- DOI: 10.3389/fphys.2023.1151495
Pericyte-derived cells participate in optic nerve scar formation
Abstract
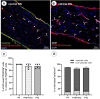
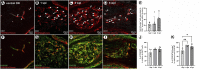
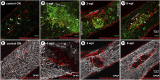
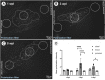
Introduction: Pericytes (PCs) are specialized cells located abluminal of endothelial cells on capillaries, fulfilling numerous important functions. Their potential involvement in wound healing and scar formation is achieving increasing attention since years. Thus, many studies investigated the participation of PCs following brain and spinal cord (SC) injury, however, lacking in-depth analysis of lesioned optic nerve (ON) tissue. Further, due to the lack of a unique PC marker and uniform definition of PCs, contradicting results are published. Methods: In the present study the inducible PDGFRβ-P2A-CreERT2-tdTomato lineage tracing reporter mouse was used to investigate the participation and trans-differentiation of endogenous PC-derived cells in an ON crush (ONC) injury model, analyzing five different post lesion time points up to 8 weeks post lesion. Results: PC-specific labeling of the reporter was evaluated and confirmed in the unlesioned ON of the reporter mouse. After ONC, we detected PC-derived tdTomato+ cells in the lesion, whereof the majority is not associated with vascular structures. The number of PC-derived tdTomato+ cells within the lesion increased over time, accounting for 60-90% of all PDGFRβ+ cells in the lesion. The presence of PDGFRβ+tdTomato- cells in the ON scar suggests the existence of fibrotic cell subpopulations of different origins. Discussion: Our results clearly demonstrate the presence of non-vascular associated tdTomato+ cells in the lesion core, indicating the participation of PC-derived cells in fibrotic scar formation following ONC. Thus, these PC-derived cells represent promising target cells for therapeutic treatment strategies to modulate fibrotic scar formation to improve axonal regeneration.
Keywords: fibrotic cells; inducible PDGFRβ-P2A-CreERT2-tdTomato lineage tracing reporter mouse; optic nerve crush; pericyte; scar formation.
Copyright © 2023 Preishuber-Pflügl, Mayr, Altinger, Brunner, Koller, Runge, Ladek, Lenzhofer, Rivera, Tempfer, Aigner, Reitsamer and Trost.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bruckner D., Kaser-Eichberger A., Bogner B., Runge C., Schrodl F., Strohmaier C., et al. (2018). Retinal pericytes: Characterization of vascular development-dependent induction time points in an inducible NG2 reporter mouse model. Curr. Eye Res. 43 (10), 1274–1285. 10.1080/02713683.2018.1493130 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

