Nucleotide excision repair in Human cell lines lacking both XPC and CSB proteins
- PMID: 37144462
- PMCID: PMC10325923
- DOI: 10.1093/nar/gkad334
Nucleotide excision repair in Human cell lines lacking both XPC and CSB proteins
Abstract
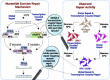
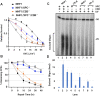
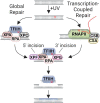
Nucleotide excision repair removes UV-induced DNA damage through two distinct sub-pathways, global repair and transcription-coupled repair (TCR). Numerous studies have shown that in human and other mammalian cell lines that the XPC protein is required for repair of DNA damage from nontranscribed DNA via global repair and the CSB protein is required for repair of lesions from transcribed DNA via TCR. Therefore, it is generally assumed that abrogating both sub-pathways with an XPC-/-/CSB-/- double mutant would eliminate all nucleotide excision repair. Here we describe the construction of three different XPC-/-/CSB-/- human cell lines that, contrary to expectations, perform TCR. The XPC and CSB genes were mutated in cell lines derived from Xeroderma Pigmentosum patients as well as from normal human fibroblasts and repair was analyzed at the whole genome level using the very sensitive XR-seq method. As predicted, XPC-/- cells exhibited only TCR and CSB-/- cells exhibited only global repair. However, the XPC-/-/CSB-/- double mutant cell lines, although having greatly reduced repair, exhibited TCR. Mutating the CSA gene to generate a triple mutant XPC-/-/CSB-/-/CSA-/- cell line eliminated all residual TCR activity. Together, these findings provide new insights into the mechanistic features of mammalian nucleotide excision repair.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
CSB-independent, XPC-dependent transcription-coupled repair in Drosophila.Proc Natl Acad Sci U S A. 2022 Mar 1;119(9):e2123163119. doi: 10.1073/pnas.2123163119. Proc Natl Acad Sci U S A. 2022. PMID: 35217627 Free PMC article.
-
Comparative analyses of two primate species diverged by more than 60 million years show different rates but similar distribution of genome-wide UV repair events.BMC Genomics. 2021 Aug 6;22(1):600. doi: 10.1186/s12864-021-07898-3. BMC Genomics. 2021. PMID: 34362292 Free PMC article.
-
XPC-RAD23B enhances UV-DDB binding to DNA to facilitate lesion search in nucleotide excision repair.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf463. doi: 10.1093/nar/gkaf463. Nucleic Acids Res. 2025. PMID: 40530698 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
UV damage induces production of mitochondrial DNA fragments with specific length profiles.Genetics. 2024 Jul 8;227(3):iyae070. doi: 10.1093/genetics/iyae070. Genetics. 2024. PMID: 38722894 Free PMC article.
-
UV damage induces production of mitochondrial DNA fragments with specific length profiles.bioRxiv [Preprint]. 2023 Nov 11:2023.11.07.566130. doi: 10.1101/2023.11.07.566130. bioRxiv. 2023. Update in: Genetics. 2024 Jul 8;227(3):iyae070. doi: 10.1093/genetics/iyae070. PMID: 37986892 Free PMC article. Updated. Preprint.
-
XPD stalled on cross-linked DNA provides insight into damage verification.Nat Struct Mol Biol. 2024 Oct;31(10):1580-1588. doi: 10.1038/s41594-024-01323-5. Epub 2024 May 28. Nat Struct Mol Biol. 2024. PMID: 38806694 Free PMC article.
-
Cross-species investigation into the requirement of XPA for nucleotide excision repair.Nucleic Acids Res. 2024 Jan 25;52(2):677-689. doi: 10.1093/nar/gkad1104. Nucleic Acids Res. 2024. PMID: 37994737 Free PMC article.
-
Genome-wide analysis of transcription-coupled repair reveals novel transcription events in Caenorhabditis elegans.PLoS Genet. 2024 Jul 19;20(7):e1011365. doi: 10.1371/journal.pgen.1011365. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 39028758 Free PMC article.
References
-
- Cleaver J.E. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968; 218:652–656. - PubMed
-
- Cleaver J.E., Bootsma D.. Xeroderma pigmentosum: biochemical and genetic characteristics. Annu. Rev. Genet. 1975; 9:19–38. - PubMed
-
- Mayne L.V., Lehmann A.R.. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982; 42:1473–1478. - PubMed
-
- Henning K.A., Li L., Iyer N., McDaniel L.D., Reagan M.S., Legerski R., Schultz R.A., Stefanini M., Lehmann A.R., Mayne L.V.et al. .. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995; 82:555–564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

