Redefining normal breast cell populations using long noncoding RNAs
- PMID: 37144467
- PMCID: PMC10325898
- DOI: 10.1093/nar/gkad339
Redefining normal breast cell populations using long noncoding RNAs
Abstract
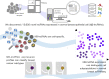
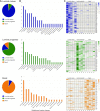
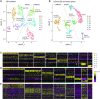
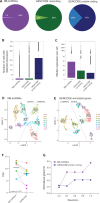
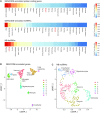
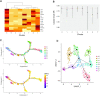
Single-cell RNAseq has allowed unprecedented insight into gene expression across different cell populations in normal tissue and disease states. However, almost all studies rely on annotated gene sets to capture gene expression levels and sequencing reads that do not align to known genes are discarded. Here, we discover thousands of long noncoding RNAs (lncRNAs) expressed in human mammary epithelial cells and analyze their expression in individual cells of the normal breast. We show that lncRNA expression alone can discriminate between luminal and basal cell types and define subpopulations of both compartments. Clustering cells based on lncRNA expression identified additional basal subpopulations, compared to clustering based on annotated gene expression, suggesting that lncRNAs can provide an additional layer of information to better distinguish breast cell subpopulations. In contrast, these breast-specific lncRNAs poorly distinguish brain cell populations, highlighting the need to annotate tissue-specific lncRNAs prior to expression analyses. We also identified a panel of 100 breast lncRNAs that could discern breast cancer subtypes better than protein-coding markers. Overall, our results suggest that lncRNAs are an unexplored resource for new biomarker and therapeutic target discovery in the normal breast and breast cancer subtypes.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Portraying breast cancers with long noncoding RNAs.Sci Adv. 2016 Sep 2;2(9):e1600220. doi: 10.1126/sciadv.1600220. eCollection 2016 Sep. Sci Adv. 2016. PMID: 27617288 Free PMC article.
-
Selective expression of long non-coding RNAs in a breast cancer cell progression model.J Cell Physiol. 2018 Feb;233(2):1291-1299. doi: 10.1002/jcp.25997. Epub 2017 Jun 15. J Cell Physiol. 2018. PMID: 28488769 Free PMC article.
-
Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia.J Biol Chem. 2015 Mar 13;290(11):6857-67. doi: 10.1074/jbc.M114.610915. Epub 2015 Jan 20. J Biol Chem. 2015. PMID: 25605728 Free PMC article.
-
Long noncoding RNAs in regulation of human breast cancer.Brief Funct Genomics. 2016 May;15(3):222-6. doi: 10.1093/bfgp/elv049. Epub 2015 Nov 18. Brief Funct Genomics. 2016. PMID: 26582840 Review.
-
Long non-coding RNAs differential expression in breast cancer subtypes: What do we know?Clin Genet. 2019 May;95(5):558-568. doi: 10.1111/cge.13502. Epub 2019 Jan 22. Clin Genet. 2019. PMID: 30614523 Review.
Cited by
-
The roles of long non-coding RNAs in ovarian cancer: from functions to therapeutic implications.Front Oncol. 2024 Apr 25;14:1332528. doi: 10.3389/fonc.2024.1332528. eCollection 2024. Front Oncol. 2024. PMID: 38725621 Free PMC article. Review.
-
Uncovering functional lncRNAs by scRNA-seq with ELATUS.Nat Commun. 2024 Nov 9;15(1):9709. doi: 10.1038/s41467-024-54005-7. Nat Commun. 2024. PMID: 39521797 Free PMC article.
-
GWAS and 3D chromatin mapping identifies multicancer risk genes associated with hormone-dependent cancers.PLoS Genet. 2024 Nov 25;20(11):e1011490. doi: 10.1371/journal.pgen.1011490. eCollection 2024 Nov. PLoS Genet. 2024. PMID: 39585897 Free PMC article.
-
CRISPR-Cas13d screens identify KILR, a breast cancer risk-associated lncRNA that regulates DNA replication and repair.Mol Cancer. 2024 May 15;23(1):101. doi: 10.1186/s12943-024-02021-y. Mol Cancer. 2024. PMID: 38745269 Free PMC article.
References
-
- Fu N.Y., Nolan E., Lindeman G.J., Visvader J.E.. Stem cells and the differentiation hierarchy in mammary gland development. Physiol. Rev. 2020; 100:489–523. - PubMed
-
- Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E.. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014; 506:322–327. - PubMed
-
- Lloyd-Lewis B., Harris O.B., Watson C.J., Davis F.M.. Mammary stem cells: premise, properties, and perspectives. Trends Cell Biol. 2017; 27:556–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

