The long and short of EJC-independent nonsense-mediated RNA decay
- PMID: 37145092
- PMCID: PMC12135299
- DOI: 10.1042/BST20221131
The long and short of EJC-independent nonsense-mediated RNA decay
Abstract
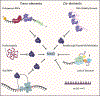
Nonsense-mediated RNA decay (NMD) plays a dual role as an RNA surveillance mechanism against aberrant transcripts containing premature termination codons and as a gene regulatory mechanism for normal physiological transcripts. This dual function is possible because NMD recognizes its substrates based on the functional definition of a premature translation termination event. An efficient mode of NMD target recognition involves the presence of exon-junction complexes (EJCs) downstream of the terminating ribosome. A less efficient, but highly conserved, mode of NMD is triggered by long 3' untranslated regions (UTRs) that lack EJCs (termed EJC-independent NMD). While EJC-independent NMD plays an important regulatory role across organisms, our understanding of its mechanism, especially in mammalian cells, is incomplete. This review focuses on EJC-independent NMD and discusses the current state of knowledge and factors that contribute to the variability in the efficiency of this mechanism.
Keywords: RNA decay; alternative splicing; exon-junction complexes; nonsense-mediated decay; translation.
© 2023 The Author(s). Published by Portland Press Limited on behalf of the Biochemical Society.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

