ONC201 in combination with paxalisib for the treatment of H3K27-altered diffuse midline glioma
- PMID: 37145169
- PMCID: PMC10345962
- DOI: 10.1158/0008-5472.CAN-23-0186
ONC201 in combination with paxalisib for the treatment of H3K27-altered diffuse midline glioma
Abstract
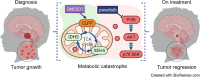
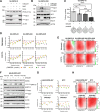
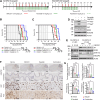
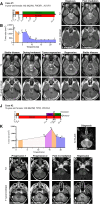
Diffuse midline gliomas (DMG), including diffuse intrinsic pontine gliomas (DIPGs), are the most lethal of childhood cancers. Palliative radiotherapy is the only established treatment, with median patient survival of 9-11 months. ONC201 is a DRD2 antagonist and ClpP agonist that has shown preclinical and emerging clinical efficacy in DMG. However, further work is needed to identify the mechanisms of response of DIPGs to ONC201 treatment and to determine whether recurring genomic features influence response. Using a systems-biological approach, we showed that ONC201 elicits potent agonism of the mitochondrial protease ClpP to drive proteolysis of electron transport chain and tricarboxylic acid cycle proteins. DIPGs harboring PIK3CA-mutations showed increased sensitivity to ONC201, while those harboring TP53-mutations were more resistant. Metabolic adaptation and reduced sensitivity to ONC201 was promoted by redox-activated PI3K/Akt signaling, which could be counteracted using the brain penetrant PI3K/Akt inhibitor, paxalisib. Together, these discoveries coupled with the powerful anti-DIPG/DMG pharmacokinetic and pharmacodynamic properties of ONC201 and paxalisib have provided the rationale for the ongoing DIPG/DMG phase II combination clinical trial NCT05009992.
Figures



References
-
- Hoffman LM, Veldhuijzen van Zanten SEM, Colditz N, Baugh J, Chaney B, Hoffmann M, et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): a collaborative report from the international and European society for pediatric oncology DIPG registries. J Clin Oncol 2018;36:1963–72. - PMC - PubMed
-
- Pincus DW, Richter EO, Yachnis AT, Bennett J, Bhatti MT, Smith A. Brainstem stereotactic biopsy sampling in children. J Neurosurg 2006;104:108–14. - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

