Reduction of Paraoxonase Expression Followed by Inactivation across Independent Semiaquatic Mammals Suggests Stepwise Path to Pseudogenization
- PMID: 37146172
- PMCID: PMC10202596
- DOI: 10.1093/molbev/msad104
Reduction of Paraoxonase Expression Followed by Inactivation across Independent Semiaquatic Mammals Suggests Stepwise Path to Pseudogenization
Abstract
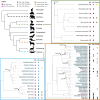
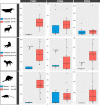
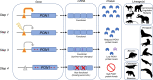
Convergent adaptation to the same environment by multiple lineages frequently involves rapid evolutionary change at the same genes, implicating these genes as important for environmental adaptation. Such adaptive molecular changes may yield either change or loss of protein function; loss of function can eliminate newly deleterious proteins or reduce energy necessary for protein production. We previously found a striking case of recurrent pseudogenization of the Paraoxonase 1 (Pon1) gene among aquatic mammal lineages-Pon1 became a pseudogene with genetic lesions, such as stop codons and frameshifts, at least four times independently in aquatic and semiaquatic mammals. Here, we assess the landscape and pace of pseudogenization by studying Pon1 sequences, expression levels, and enzymatic activity across four aquatic and semiaquatic mammal lineages: pinnipeds, cetaceans, otters, and beavers. We observe in beavers and pinnipeds an unexpected reduction in expression of Pon3, a paralog with similar expression patterns but different substrate preferences. Ultimately, in all lineages with aquatic/semiaquatic members, we find that preceding any coding-level pseudogenization events in Pon1, there is a drastic decrease in expression, followed by relaxed selection, thus allowing accumulation of disrupting mutations. The recurrent loss of Pon1 function in aquatic/semiaquatic lineages is consistent with a benefit to Pon1 functional loss in aquatic environments. Accordingly, we examine diving and dietary traits across pinniped species as potential driving forces of Pon1 functional loss. We find that loss is best associated with diving activity and likely results from changes in selective pressures associated with hypoxia and hypoxia-induced inflammation.
Keywords: convergent evolution; genomics; hypoxia; marine; pseudogene.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


References
-
- Aharoni S, Aviram M, Fuhrman B. 2013. Paraoxonase 1 (PON1) reduces macrophage inflammatory responses. Atherosclerosis. 228:353–361. - PubMed
-
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet. 17:379. - PubMed
-
- Balakirev ES, Ayala FJ. 2003. Pseudogenes: are they “junk” or functional DNA? Annu Rev Genet. 37:123–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

