The functional impact of 1,570 individual amino acid substitutions in human OTC
- PMID: 37146589
- PMCID: PMC10183466
- DOI: 10.1016/j.ajhg.2023.03.019
The functional impact of 1,570 individual amino acid substitutions in human OTC
Abstract
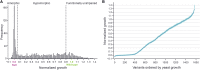
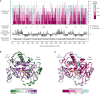
Deleterious mutations in the X-linked gene encoding ornithine transcarbamylase (OTC) cause the most common urea cycle disorder, OTC deficiency. This rare but highly actionable disease can present with severe neonatal onset in males or with later onset in either sex. Individuals with neonatal onset appear normal at birth but rapidly develop hyperammonemia, which can progress to cerebral edema, coma, and death, outcomes ameliorated by rapid diagnosis and treatment. Here, we develop a high-throughput functional assay for human OTC and individually measure the impact of 1,570 variants, 84% of all SNV-accessible missense mutations. Comparison to existing clinical significance calls, demonstrated that our assay distinguishes known benign from pathogenic variants and variants with neonatal onset from late-onset disease presentation. This functional stratification allowed us to identify score ranges corresponding to clinically relevant levels of impairment of OTC activity. Examining the results of our assay in the context of protein structure further allowed us to identify a 13 amino acid domain, the SMG loop, whose function appears to be required in human cells but not in yeast. Finally, inclusion of our data as PS3 evidence under the current ACMG guidelines, in a pilot reclassification of 34 variants with complete loss of activity, would change the classification of 22 from variants of unknown significance to clinically actionable likely pathogenic variants. These results illustrate how large-scale functional assays are especially powerful when applied to rare genetic diseases.
Keywords: OTC deficiency; Oxford Nanopore sequencing; SNV analysis/discovery; X-linked disease; metabolic disorder; model organisms; multiplexed assays of variant effect; rare disease; rare variants; urea cycle disorder; variant interpretation.
Copyright © 2023 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.M.D. is a scientific advisor with a financial interest in Fenologica Biosciences, Inc. H.M. is a founding member of Cogthera LLC. A.M.D., R.S.L., G.A.C., A.S., and L.C. are listed as inventors on US patents or patent applications that may be relevant to this work.
Figures






References
-
- Summar M.L., Koelker S., Freedenberg D., Le Mons C., Haberle J., Lee H.S., Kirmse B., European Registry and Network for Intoxication Type Metabolic Diseases. Members of the Urea Cycle Disorders Consortium UCDC The incidence of urea cycle disorders. Mol. Genet. Metab. 2013;110:179–180. - PMC - PubMed
-
- Ausems M.G., Bakker E., Berger R., Duran M., van Diggelen O.P., Keulemans J.L., de Valk H.W., Kneppers A.L., Dorland L., Eskes P.F., et al. Asymptomatic and late-onset ornithine transcarbamylase deficiency caused by a A208T mutation: clinical, biochemical and DNA analyses in a four-generation family. Am. J. Med. Genet. 1997;68:236–239. - PubMed
-
- van Diggelen O.P., Zaremba J., He W., Keulemans J.L., Boer A.M., Reuser A.J., Ausems M.G., Smeitink J.A., Kowalczyk J., Pronicka E., et al. Asymptomatic and late-onset ornithine transcarbamylase (OTC) deficiency in males of a five-generation family, caused by an A208T mutation. Clin. Genet. 1996;50:310–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

