KK-LC-1 as a therapeutic target to eliminate ALDH+ stem cells in triple negative breast cancer
- PMID: 37147285
- PMCID: PMC10163259
- DOI: 10.1038/s41467-023-38097-1
KK-LC-1 as a therapeutic target to eliminate ALDH+ stem cells in triple negative breast cancer
Abstract
Failure to achieve complete elimination of triple negative breast cancer (TNBC) stem cells after adjuvant therapy is associated with poor outcomes. Aldehyde dehydrogenase 1 (ALDH1) is a marker of breast cancer stem cells (BCSCs), and its enzymatic activity regulates tumor stemness. Identifying upstream targets to control ALDH+ cells may facilitate TNBC tumor suppression. Here, we show that KK-LC-1 determines the stemness of TNBC ALDH+ cells via binding with FAT1 and subsequently promoting its ubiquitination and degradation. This compromises the Hippo pathway and leads to nuclear translocation of YAP1 and ALDH1A1 transcription. These findings identify the KK-LC-1-FAT1-Hippo-ALDH1A1 pathway in TNBC ALDH+ cells as a therapeutic target. To reverse the malignancy due to KK-LC-1 expression, we employ a computational approach and discover Z839878730 (Z8) as an small-molecule inhibitor which may disrupt KK-LC-1 and FAT1 binding. We demonstrate that Z8 suppresses TNBC tumor growth via a mechanism that reactivates the Hippo pathway and decreases TNBC ALDH+ cell stemness and viability.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

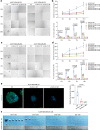
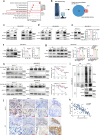
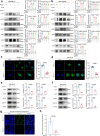

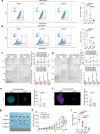

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

