The expression pattern of pyruvate dehydrogenase kinases predicts prognosis and correlates with immune exhaustion in clear cell renal cell carcinoma
- PMID: 37147361
- PMCID: PMC10162970
- DOI: 10.1038/s41598-023-34087-x
The expression pattern of pyruvate dehydrogenase kinases predicts prognosis and correlates with immune exhaustion in clear cell renal cell carcinoma
Erratum in
-
Author Correction: The expression pattern of pyruvate dehydrogenase kinases predicts prognosis and correlates with immune exhaustion in clear cell renal cell carcinoma.Sci Rep. 2023 Jun 6;13(1):9186. doi: 10.1038/s41598-023-36328-5. Sci Rep. 2023. PMID: 37280349 Free PMC article. No abstract available.
Abstract
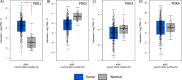
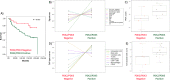
Renal cancer cells constitute a paradigm of tumor cells with a glycolytic reprogramming which drives metabolic alterations favouring cell survival and transformation. We studied the expression and activity of pyruvate dehydrogenase kinases (PDK1-4), key enzymes of the energy metabolism, in renal cancer cells. We analysed the expression, subcellular distribution and clinicopathological correlations of PDK1-4 by immunohistochemistry of tumor tissue microarray samples from a cohort of 96 clear cell renal cell carcinoma (ccRCC) patients. Gene expression analysis was performed on whole tumor tissue sections of a subset of ccRCC samples. PDK2 and PDK3 protein expression in tumor cells correlated with lower patient overall survival, whereas PDK1 protein expression correlated with higher patient survival. Gene expression analysis revealed molecular association of PDK2 and PDK3 expression with PI3K signalling pathway, as well as with T cell infiltration and exhausted CD8 T cells. Inhibition of PDK by dichloroacetate in human renal cancer cell lines resulted in lower cell viability, which was accompanied by an increase in pAKT. Together, our findings suggest a differential role for PDK enzymes in ccRCC progression, and highlight PDK as actionable metabolic proteins in relation with PI3K signalling and exhausted CD8 T cells in ccRCC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

