Anti-inflammatory effects of CBD in human microglial cell line infected with HIV-1
- PMID: 37147420
- PMCID: PMC10162654
- DOI: 10.1038/s41598-023-32927-4
Anti-inflammatory effects of CBD in human microglial cell line infected with HIV-1
Abstract
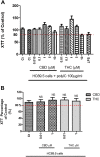
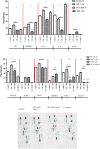
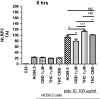
Human immunodeficiency virus (HIV) infection is associated with a chronic inflammatory stage and continuous activation of inflammasome pathway. We studied the anti-inflammatory effects of the compound cannabidiol (CBD) in comparison with Δ (9)-tetrahydrocannabinol [Δ(9)-THC] in human microglial cells (HC69.5) infected with HIV. Our results showed that CBD reduced the production of various inflammatory cytokines and chemokines such as MIF, SERPIN E1, IL-6, IL-8, GM-CSF, MCP-1, CXCL1, CXCL10, and IL-1 β compared to Δ(9)-THC treatment. In addition, CBD led to the deactivation of caspase 1, reduced NLRP3 gene expression which play a crucial role in the inflammasome cascade. Furthermore, CBD significantly reduced the expression of HIV. Our study demonstrated that CBD has anti-inflammatory properties and exhibits significant therapeutic potential against HIV-1 infections and neuroinflammation.
© 2023. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

