Environmental control of rice flowering time
- PMID: 37147799
- PMCID: PMC10504588
- DOI: 10.1016/j.xplc.2023.100610
Environmental control of rice flowering time
Abstract
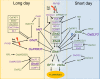
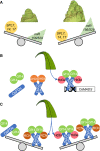
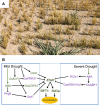
Correct measurement of environmental parameters is fundamental for plant fitness and survival, as well as for timing developmental transitions, including the switch from vegetative to reproductive growth. Important parameters that affect flowering time include day length (photoperiod) and temperature. Their response pathways have been best described in Arabidopsis, which currently offers a detailed conceptual framework and serves as a comparison for other species. Rice, the focus of this review, also possesses a photoperiodic flowering pathway, but 150 million years of divergent evolution in very different environments have diversified its molecular architecture. The ambient temperature perception pathway is strongly intertwined with the photoperiod pathway and essentially converges on the same genes to modify flowering time. When observing network topologies, it is evident that the rice flowering network is centered on EARLY HEADING DATE 1, a rice-specific transcriptional regulator. Here, we summarize the most important features of the rice photoperiodic flowering network, with an emphasis on its uniqueness, and discuss its connections with hormonal, temperature perception, and stress pathways.
Keywords: florigens; flowering; photoperiod; rice; stress; temperature.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Abe M., Kosaka S., Shibuta M., Nagata K., Uemura T., Nakano A., Kaya H. Transient activity of the florigen complex during the floral transition in arabidopsis thaliana. Devenir. 2019;146 - PubMed
-
- Andrés F., Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. - PubMed

