Immune mechanisms shape the clonal landscape during early progression of prostate cancer
- PMID: 37148881
- PMCID: PMC10330387
- DOI: 10.1016/j.devcel.2023.04.010
Immune mechanisms shape the clonal landscape during early progression of prostate cancer
Abstract
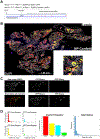
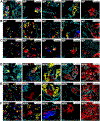
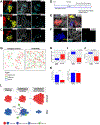
Understanding the role of the immune microenvironment in modulating intratumor heterogeneity is essential for effective cancer therapies. Using multicolor lineage tracing in genetically engineered mouse models and single-cell transcriptomics, we show that slowly progressing tumors contain a multiclonal landscape of relatively homogeneous subpopulations within a well-organized tumor microenvironment. In more advanced and aggressive tumors, however, the multiclonal landscape develops into competing dominant and minor clones accompanied by a disordered microenvironment. We demonstrate that this dominant/minor landscape is associated with differential immunoediting, in which minor clones are marked by an increased expression of IFNγ-response genes and the T cell-activating chemokines Cxcl9 and Cxcl11. Furthermore, immunomodulation of the IFNγ pathway can rescue minor clones from elimination. Notably, the immune-specific gene signature of minor clones exhibits a prognostic value for biochemical recurrence-free survival in human prostate cancer. These findings suggest new immunotherapy approaches for modulating clonal fitness and tumor progression in prostate cancer.
Keywords: clonal competition; immunoediting; prostate cancer; single-cell transcriptomics.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

