Genome-wide identification and expression profile analysis of metal tolerance protein gene family in Eucalyptus grandis under metal stresses
- PMID: 37149585
- PMCID: PMC10163719
- DOI: 10.1186/s12870-023-04240-9
Genome-wide identification and expression profile analysis of metal tolerance protein gene family in Eucalyptus grandis under metal stresses
Abstract
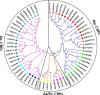
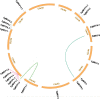
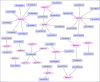
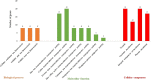
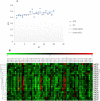
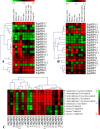
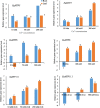
Metal tolerance proteins (MTPs) as Me2+/H+(K+) antiporters participate in the transport of divalent cations, leading to heavy metal stress resistance and mineral utilization in plants. In the present study, to obtain better knowledge of the biological functions of the MTPs family, 20 potential EgMTPs genes were identified in Eucalyptus grandis and classified into seven groups belonging to three cation diffusion facilitator groups (Mn-CDFs, Zn/Fe-CDFs, and Zn-CDFs) and seven groups. EgMTP-encoded amino acids ranged from 315 to 884, and most of them contained 4-6 recognized transmembrane domains and were clearly prognosticated to localize into the cell vacuole. Almost all EgMTP genes experienced gene duplication events, in which some might be uniformly distributed in the genome. The numbers of cation efflux and the zinc transporter dimerization domain were highest in EgMTP proteins. The promoter regions of EgMTP genes have different cis-regulatory elements, indicating that the transcription rate of EgMTP genes can be a controlled response to different stimuli in multiple pathways. Our findings provide accurate perception on the role of the predicted miRNAs and the presence of SSR marker in the Eucalyptus genome and clarify their functions in metal tolerance regulation and marker-assisted selection, respectively. Gene expression profiling based on previous RNA-seq data indicates a probable function for EgMTP genes during development and responses to biotic stress. Additionally, the upregulation of EgMTP6, EgMTP5, and EgMTP11.1 to excess Cd2+ and Cu2+ exposure might be responsible for metal translocation from roots to leaves.
Keywords: Eucalyptus grandis; Expression profile; Genome-wide identification; Heavy metals; Metal tolerance protein (MTP).
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genome-wide identification and expression analysis of metal tolerance protein (MTP) gene family in soybean (Glycine max) under heavy metal stress.Mol Biol Rep. 2023 Apr;50(4):2975-2990. doi: 10.1007/s11033-022-08100-x. Epub 2023 Jan 18. Mol Biol Rep. 2023. PMID: 36653731
-
Genome-Wide Identification and Expression Analysis of Metal Tolerance Protein Gene Family in Medicago truncatula Under a Broad Range of Heavy Metal Stress.Front Genet. 2021 Sep 7;12:713224. doi: 10.3389/fgene.2021.713224. eCollection 2021. Front Genet. 2021. PMID: 34603378 Free PMC article.
-
Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): insights into metal homeostasis and biofortification.Biometals. 2017 Apr;30(2):217-235. doi: 10.1007/s10534-017-9997-x. Epub 2017 Feb 1. Biometals. 2017. PMID: 28150142
-
Cation Diffusion Facilitator family: Structure and function.FEBS Lett. 2015 May 22;589(12):1283-95. doi: 10.1016/j.febslet.2015.04.007. Epub 2015 Apr 17. FEBS Lett. 2015. PMID: 25896018 Review.
-
Emerging mechanisms for heavy metal transport in plants.Biochim Biophys Acta. 2000 May 1;1465(1-2):104-26. doi: 10.1016/s0005-2736(00)00133-4. Biochim Biophys Acta. 2000. PMID: 10748249 Review.
Cited by
-
Genome-wide identification, expression and function analysis of the MTP gene family in tulip (Tulipa gesneriana).Front Plant Sci. 2024 Feb 19;15:1346255. doi: 10.3389/fpls.2024.1346255. eCollection 2024. Front Plant Sci. 2024. PMID: 38439986 Free PMC article.
-
Organic fertilizer improved the lead and cadmium metal tolerance of Eucalyptus camaldulensis by enhancing the uptake of potassium, phosphorus, and calcium.Front Plant Sci. 2024 Sep 23;15:1444227. doi: 10.3389/fpls.2024.1444227. eCollection 2024. Front Plant Sci. 2024. PMID: 39376235 Free PMC article.
-
Synergistic interactions of assorted ameliorating agents to enhance the potential of heavy metal phytoremediation.Stress Biol. 2024 Feb 16;4(1):13. doi: 10.1007/s44154-024-00153-1. Stress Biol. 2024. PMID: 38363436 Free PMC article. Review.
References
-
- Marschner H. Marschner's mineral nutrition of higher plants. 3. United States: Academic; 2011.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

