Roles of adenine methylation in the physiology of Lacticaseibacillus paracasei
- PMID: 37149616
- PMCID: PMC10164179
- DOI: 10.1038/s41467-023-38291-1
Roles of adenine methylation in the physiology of Lacticaseibacillus paracasei
Abstract
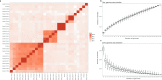
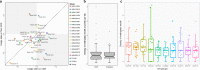
Lacticaseibacillus paracasei is an economically important bacterial species, used in the food industry and as a probiotic. Here, we investigate the roles of N6-methyladenine (6mA) modification in L. paracasei using multi-omics and high-throughput chromosome conformation capture (Hi-C) analyses. The distribution of 6mA-modified sites varies across the genomes of 28 strains, and appears to be enriched near genes involved in carbohydrate metabolism. A pglX mutant, defective in 6mA modification, shows transcriptomic alterations but only modest changes in growth and genomic spatial organization.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Vandenbussche, I. et al. Detection of cytosine methylation in Burkholderia cenocepacia by single-molecule real-time sequencing and whole-genome bisulfite sequencing. Microbiology (Reading) 167 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

