Formalin-fixed paraffin-embedded (FFPE) samples help to investigate transcriptomic responses in wildlife disease
- PMID: 37150904
- PMCID: PMC12142723
- DOI: 10.1111/1755-0998.13805
Formalin-fixed paraffin-embedded (FFPE) samples help to investigate transcriptomic responses in wildlife disease
Abstract
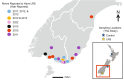
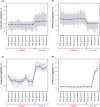
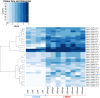
Infectious diseases impact numerous organisms. Knowledge of host-pathogen interactions and host responses to infection is crucial for conservation and management. Obtaining this knowledge quickly is made increasingly possible by a variety of genomic approaches, yet, for many species the bottleneck to understanding this, remains access to appropriate samples and data. Lack of sample availability has also limited our understanding of how pathogens and the immune responses of hosts change over time. Archival materials may provide a way to explore pathogen emergence and host responses over multiple-possibly hundreds-of years. Here, we tested whether formalin-fixed paraffin-embedded (FFPE) tissue samples could be used to understand an unknown pathology, lamprey reddening syndrome (LRS), affecting pouched lampreys (Geotria australis). Our differential expression analyses of dermal tissues from four unaffected lampreys and eight affected lampreys collected in 2012 alluded to several potential agents associated with LRS. Interestingly, the pathways associated with viral infections were overrepresented in affected versus unaffected lamprey. Gene ontology analyses of the affected and non-affected lampreys also provided new insights into the largely understudied immune responses of pouched lampreys. Our work confirms that FFPE samples can be used to infer information about the transcriptional responses of a wildlife species affected by unknown historical pathologies/syndromes. In addition, the use of FFPE samples for transcriptomics offers many opportunities to investigate the genomic responses of a species to a variety of environmental changes. We conclude with a discussion about how to best sample and utilize these unique archival resources for future wildlife transcriptomic studies.
Keywords: conservation; fish; historical pathologies; immune response; lamprey reddening syndrome; pouched lamprey.
© 2023 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures




Similar articles
-
Preparation of archival formalin-fixed paraffin-embedded mouse liver samples for use with the Agilent gene expression microarray platform.J Pharmacol Toxicol Methods. 2013 Sep-Oct;68(2):260-268. doi: 10.1016/j.vascn.2013.02.008. Epub 2013 Mar 1. J Pharmacol Toxicol Methods. 2013. PMID: 23458726
-
Editor's Highlight: Dose-Response Analysis of RNA-Seq Profiles in Archival Formalin-Fixed Paraffin-Embedded Samples.Toxicol Sci. 2016 Dec;154(2):202-213. doi: 10.1093/toxsci/kfw161. Epub 2016 Aug 25. Toxicol Sci. 2016. PMID: 27562560
-
Mining the Archives: A Cross-Platform Analysis of Gene Expression Profiles in Archival Formalin-Fixed Paraffin-Embedded Tissues.Toxicol Sci. 2015 Dec;148(2):460-72. doi: 10.1093/toxsci/kfv195. Epub 2015 Sep 10. Toxicol Sci. 2015. PMID: 26361796 Free PMC article.
-
Challenges and solutions in FISH for formalin-fixed paraffin-embedded tissue: A scoping review.Microsc Res Tech. 2025 Jan;88(1):270-278. doi: 10.1002/jemt.24702. Epub 2024 Sep 24. Microsc Res Tech. 2025. PMID: 39315587
-
Systematic review and feasibility study on pre-analytical factors and genomic analyses on archival formalin-fixed paraffin-embedded breast cancer tissue.Sci Rep. 2024 Aug 6;14(1):18275. doi: 10.1038/s41598-024-69285-8. Sci Rep. 2024. PMID: 39107471 Free PMC article.
Cited by
-
Screening great ape museum specimens for DNA viruses.Sci Rep. 2024 Nov 30;14(1):29806. doi: 10.1038/s41598-024-80780-w. Sci Rep. 2024. PMID: 39616255 Free PMC article.
References
-
- Adlard, R. D. , Miller, T. L. , & Smit, N. J. (2015). The butterfly effect: Parasite diversity, environment, and emerging disease in aquatic wildlife. Trends in Parasitology, 31(4), 160–166. - PubMed
-
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. - PubMed
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute.
-
- Arreaza, G. , Qiu, P. , Pang, L. , Albright, A. , Hong, L. Z. , Marton, M. J. , & Levitan, D. (2016). Pre‐analytical considerations for successful next‐generation sequencing (NGS): Challenges and opportunities for formalin‐fixed and paraffin‐embedded tumor tissue (FFPE) samples. International Journal of Molecular Sciences, 17(9), 1579. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

