De novo transcriptome sequencing and gene co-expression reveal a genomic basis for drought sensitivity and evidence of a rapid local adaptation on Atlas cedar (Cedrus atlantica)
- PMID: 37152146
- PMCID: PMC10155838
- DOI: 10.3389/fpls.2023.1116863
De novo transcriptome sequencing and gene co-expression reveal a genomic basis for drought sensitivity and evidence of a rapid local adaptation on Atlas cedar (Cedrus atlantica)
Abstract
Introduction: Understanding the adaptive capacity to current climate change of drought-sensitive tree species is mandatory, given their limited prospect of migration and adaptation as long-lived, sessile organisms. Knowledge about the molecular and eco-physiological mechanisms that control drought resilience is thus key, since water shortage appears as one of the main abiotic factors threatening forests ecosystems. However, our current background is scarce, especially in conifers, due to their huge and complex genomes.
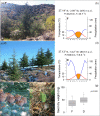
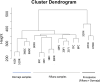
Methods: Here we investigated the eco-physiological and transcriptomic basis of drought response of the climate change-threatened conifer Cedrus atlantica. We studied C. atlantica seedlings from two locations with contrasting drought conditions to investigate a local adaptation. Seedlings were subjected to experimental drought conditions, and were monitored at immediate (24 hours) and extended (20 days) times. In addition, post-drought recovery was investigated, depicting two contrasting responses in both locations (drought resilient and non-resilient). Single nucleotide polymorphisms (SNPs) were also studied to characterize the genomic basis of drought resilience and investigate a rapid local adaptation of C. atlantica.
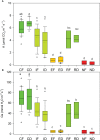
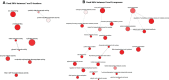
Results: De novo transcriptome assembly was performed for the first time in this species, providing differences in gene expression between the immediate and extended treatments, as well as among the post-drought recovery phenotypes. Weighted gene co-expression network analysis showed a regulation of stomatal closing and photosynthetic activity during the immediate drought, consistent with an isohydric dynamic. During the extended drought, growth and flavonoid biosynthesis inhibition mechanisms prevailed, probably to increase root-to-shoot ratio and to limit the energy-intensive biosynthesis of secondary metabolites. Drought sensitive individuals failed in metabolism and photosynthesis regulation under drought stress, and in limiting secondary metabolite production. Moreover, genomic differences (SNPs) were found between drought resilient and sensitive seedlings, and between the two studied locations, which were mostly related to transposable elements.
Discussion: This work provides novel insights into the transcriptomic basis of drought response of C. atlantica, a set of candidate genes mechanistically involved in its drought sensitivity and evidence of a rapid local adaptation. Our results may help guide conservation programs for this threatened conifer, contribute to advance drought-resilience research and shed light on trees' adaptive potential to current climate change.
Keywords: Atlas cedar; RNA-Seq; adaptive capacity; climate change; conifers; drought sensitiveness; eco-physiology; phenotypic diversity.
Copyright © 2023 Cobo-Simón, Gómez-Garrido, Esteve-Codina, Dabad, Alioto, Maloof, Méndez-Cea, Seco, Linares and Gallego.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Contrasting transcriptomic patterns reveal a genomic basis for drought resilience in the relict fir Abies pinsapo Boiss.Tree Physiol. 2023 Feb 4;43(2):315-334. doi: 10.1093/treephys/tpac115. Tree Physiol. 2023. PMID: 36210755
-
Differences in temperature sensitivity and drought recovery between natural stands and plantations of conifers are species-specific.Sci Total Environ. 2021 Nov 20;796:148930. doi: 10.1016/j.scitotenv.2021.148930. Epub 2021 Jul 16. Sci Total Environ. 2021. PMID: 34378542
-
Connecting tree-ring phenotypes, genetic associations and transcriptomics to decipher the genomic architecture of drought adaptation in a widespread conifer.Mol Ecol. 2021 Aug;30(16):3898-3917. doi: 10.1111/mec.15846. Epub 2021 Mar 6. Mol Ecol. 2021. PMID: 33586257 Free PMC article.
-
Comparative analysis of two sister Erythrophleum species (Leguminosae) reveal contrasting transcriptome-wide responses to early drought stress.Gene. 2019 Apr 30;694:50-62. doi: 10.1016/j.gene.2019.01.027. Epub 2019 Feb 1. Gene. 2019. PMID: 30716444
-
Atlas cedar response to climate-driven disturbances at its southern range edge.Environ Monit Assess. 2025 May 24;197(6):673. doi: 10.1007/s10661-025-14140-0. Environ Monit Assess. 2025. PMID: 40413346
Cited by
-
Comparative Stem Transcriptome Analysis Reveals Pathways Associated with Drought Tolerance in Maritime Pine Grafts.Int J Mol Sci. 2024 Sep 14;25(18):9926. doi: 10.3390/ijms25189926. Int J Mol Sci. 2024. PMID: 39337414 Free PMC article.
-
Maritime Pine Rootstock Genotype Modulates Gene Expression Associated with Stress Tolerance in Grafted Stems.Plants (Basel). 2024 Jun 14;13(12):1644. doi: 10.3390/plants13121644. Plants (Basel). 2024. PMID: 38931075 Free PMC article.
-
Differential gene reactions reveal drought response strategies in African acacias.Plant J. 2025 Aug;123(3):e70385. doi: 10.1111/tpj.70385. Plant J. 2025. PMID: 40758440 Free PMC article.
-
Genome-wide DNA polymorphisms in two peatland adapted Coffea liberica varieties.BMC Genom Data. 2025 Feb 14;26(1):11. doi: 10.1186/s12863-025-01305-6. BMC Genom Data. 2025. PMID: 39953379 Free PMC article.
-
The transcriptome sequencing analysis reveals immune mechanisms of soybean fermented powder on the loach (Misgurnus anguillicaudatus) in response to Lipopolysaccharide (LPS) infection.Front Immunol. 2023 Aug 18;14:1247038. doi: 10.3389/fimmu.2023.1247038. eCollection 2023. Front Immunol. 2023. PMID: 37662918 Free PMC article.
References
-
- Abel-Schaad D., Iriarte E., López-Sáez J. A., Pérez-Díaz S., Sabariego Ruiz S., Cheddadi R., et al. . (2018). Are Cedrus atlantica forests in the rif mountains of Morocco heading towards local extinction? Holocene 28 (6), 1023–1037. doi: 10.1177/0959683617752842 - DOI
-
- Alba-Sánchez F., Abel-Schaad D., López-Sáez J. A., Sabariego R., Pérez-Díaz S., González-Hernández A. (2018). Paleobiogeografía de abies spp. y Cedrus atlantica en el mediterráneo occidental (península ibérica y marruecos). Ecosistemas 27 (1), 26–37. doi: 10.7818/ECOS.1441 - DOI

