Resistance screening and in silico characterization of cloned novel RGA from multi-race resistant lentil germplasm against Fusarium wilt (Fusarium oxysporum f. sp. lentis)
- PMID: 37152180
- PMCID: PMC10160667
- DOI: 10.3389/fpls.2023.1147220
Resistance screening and in silico characterization of cloned novel RGA from multi-race resistant lentil germplasm against Fusarium wilt (Fusarium oxysporum f. sp. lentis)
Abstract
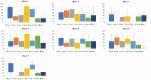
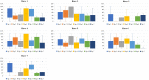
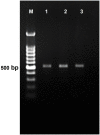
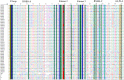
Fusarium wilt caused by Fusarium oxysporum f. sp. lentis (Fol) is the most devastating disease of lentil present worldwide. Identification of multi-race fusarium wilt resistance genes and their incorporation into existing cultivars will help to reduce yield losses. In the present study, 100 lentil germplasms belonging to seven lentil species were screened against seven prevalent races of Fol, and accessions IC201561 (Lens culinaris subsp. culinaris), EC714243 (L. c. subsp. odemensis), and EC718238 (L. nigricans) were identified as resistant. The typical R gene codes for the nucleotide-binding site and leucine-rich repeats (NBS-LRR) at the C terminal are linked to either the Toll/interleukin 1-like receptor (TIR) or coiled coil (CC) at the N terminal. In the present study, degenerate primers, designed from the NBS region amplifying the P-loop to the GLPLA motif, isolated forty-five resistance gene analogues (RGAs) from identified resistant accessions. The sequence alignment identified both classes of RGAs, TIR and non-TIR, based on the presence of aspartate (D) and tryptophan (W) at the end of the kinase motif, respectively. The phylogenetic analysis grouped the RGAs into six classes, from LRGA1 to LRGA6, which determined the diversity of the RGAs present in the host. Grouping of the RGAs identified from Lens nigricans, LnRGA 2, 9, 13 with I2 revealed the structural similarity with the fusarium resistance gene. The similarity index ranged from 27.85% to 86.98% among the RGAs and from 26.83% to 49.41% among the known R genes, I2, Gpa2, M, and L6. The active binding sites present along the conserved motifs grouped the RGAs into 13 groups. ADP/ATP, being the potential ligand, determines the ATP binding and ATP hydrolysis activity of the RGAs. The isolated RGAs can be used to develop markers linked to the functional R gene. Furthermore, expression analysis and full-length gene isolation pave the path to identifying the molecular mechanism involved in resistance.
Keywords: cloning; fusarium wilt; in silico characterization; lentil; resistance gene analogue; screening.
Copyright © 2023 Nishmitha, Singh, Dubey, Akthar, Tripathi and Kamil.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Expression profiling and characterization of key RGA involved in lentil Fusarium wilt Race 5 resistance.World J Microbiol Biotechnol. 2023 Sep 15;39(11):306. doi: 10.1007/s11274-023-03748-4. World J Microbiol Biotechnol. 2023. PMID: 37713019
-
Isolation and characterization of fusarium wilt resistance gene analogs in radish.3 Biotech. 2018 May;8(5):255. doi: 10.1007/s13205-018-1279-y. Epub 2018 May 14. 3 Biotech. 2018. PMID: 29765813 Free PMC article.
-
Isolation of TIR and non-TIR NBS--LRR resistance gene analogues and identification of molecular markers linked to a powdery mildew resistance locus in chestnut rose (Rosa roxburghii Tratt).Theor Appl Genet. 2005 Sep;111(5):819-30. doi: 10.1007/s00122-005-0002-7. Epub 2005 Oct 18. Theor Appl Genet. 2005. PMID: 16075209
-
Identification and expression profiling analysis of NBS-LRR genes involved in Fusarium oxysporum f.sp. conglutinans resistance in cabbage.3 Biotech. 2019 May;9(5):202. doi: 10.1007/s13205-019-1714-8. Epub 2019 May 4. 3 Biotech. 2019. PMID: 31065502 Free PMC article. Review.
-
Disease Resistance Gene Analogs (RGAs) in Plants.Int J Mol Sci. 2015 Aug 14;16(8):19248-90. doi: 10.3390/ijms160819248. Int J Mol Sci. 2015. PMID: 26287177 Free PMC article. Review.
Cited by
-
Molecular characterization of Indian races of Fusarium oxysporum f. sp. lentis (Fol) based on secreted in Xylem (SIX) effector genes and development of a SIX11 gene-based molecular marker for specific detection of Fol.Arch Microbiol. 2024 Apr 2;206(4):200. doi: 10.1007/s00203-024-03945-1. Arch Microbiol. 2024. PMID: 38564016
References
-
- Agbagwa I. O., Patil P. G., Das A., Soren K. R., Singh I. P., Chaturvedi S. K., et al. . (2018). Identification, characterization, and phylogenetic analysis of pigeonpea (Cajanus cajan l. millsp.) resistance gene analogs using PCR cloning and in silico methods. Genet. Mol. Res. 17, 1–14. doi: 10.4238/gmr16039895 - DOI
-
- Agrawal S. C., Singh K., Lal S. S. (1993). “Plant protection of lentil in India,” in Lentil in south Asia. Eds. Erskine W., Saxena M. C. (Aleppo, Syria: ICARDA; ), pp147–pp165.
-
- Ahmed S., Akem C., Bayaa B., Erskine W. (2002). Integrating host resistance with planting date and fungicide seed treatment to manage fusarium wilt and so increase lentil yields. Int. J. Pest Manag 48, 121–125. doi: 10.1080/09670870110097690 - DOI
-
- Alvarez J. M., González-Torres R., Mallor C., Gómez-Guillamón M. L. (2005). Potential sources of resistance to fusarium wilt and powdery mildew in melons. HortScience. 40, 1657–1660. doi: 10.21273/HORTSCI.40.6.1657 - DOI
-
- Arumuganathan K., Earle E. D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. doi: 10.1007/BF02672069 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

