A new osteogenic protein isolated from Dioscorea opposita Thunb accelerates bone defect healing through the mTOR signaling axis
- PMID: 37152710
- PMCID: PMC10160600
- DOI: 10.1016/j.bioactmat.2023.04.018
A new osteogenic protein isolated from Dioscorea opposita Thunb accelerates bone defect healing through the mTOR signaling axis
Abstract
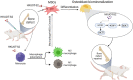
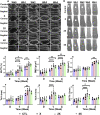
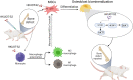
Delayed bone defect repairs lead to severe health and socioeconomic impacts on patients. Hence, there are increasing demands for medical interventions to promote bone defect healing. Recombinant proteins such as BMP-2 have been recognized as one of the powerful osteogenic substances that promote mesenchymal stem cells (MSCs) to osteoblast differentiation and are widely applied clinically for bone defect repairs. However, recent reports show that BMP-2 treatment has been associated with clinical adverse side effects such as ectopic bone formation, osteolysis and stimulation of inflammation. Here, we have identified one new osteogenic protein, named 'HKUOT-S2' protein, from Dioscorea opposita Thunb. Using the bone defect model, we have shown that the HKUOT-S2 protein can accelerate bone defect repair by activating the mTOR signaling axis of MSCs-derived osteoblasts and increasing osteoblastic biomineralization. The HKUOT-S2 protein can also modulate the transcriptomic changes of macrophages, stem cells, and osteoblasts, thereby enhancing the crosstalk between the polarized macrophages and MSCs-osteoblast differentiation to facilitate osteogenesis. Furthermore, this protein had no toxic effects in vivo. We have also identified HKUOT-S2 peptide sequence TKSSLPGQTK as a functional osteogenic unit that can promote osteoblast differentiation in vitro. The HKUOT-S2 protein with robust osteogenic activity could be a potential alternative osteoanabolic agent for promoting osteogenesis and bone defect repairs. We believe that the HKUOT-S2 protein may potentially be applied clinically as a new class of osteogenic agent for bone defect healing.
Keywords: Bone defect repair; Bone mineral density (BMD); Dioscorea spp protein; Mesenchymal stem cells (MSCs); Osteoblast differentiation; mTOR signaling pathway.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures












References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

