Efficient SARS-CoV-2 Surveillance during the Pandemic-Endemic Transition Using PCR-Based Genotyping Assays
- PMID: 37154727
- PMCID: PMC10269661
- DOI: 10.1128/spectrum.03450-22
Efficient SARS-CoV-2 Surveillance during the Pandemic-Endemic Transition Using PCR-Based Genotyping Assays
Abstract
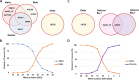
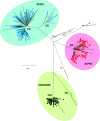
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants of concern (VOC) pose an increased risk to public health due to higher transmissibility and/or immune escape. In this study, we assessed the performance of a custom TaqMan SARS-CoV-2 mutation panel consisting of 10 selected real-time PCR (RT-PCR) genotyping assays compared to whole-genome sequencing (WGS) for identification of 5 VOC circulating in The Netherlands. SARS-CoV-2 positive samples (N = 664), collected during routine PCR screening (15 ≤ CT ≤ 32) between May-July 2021 and December 2021-January 2022, were selected and analyzed using the RT-PCR genotyping assays. VOC lineage was determined based on the detected mutation profile. In parallel, all samples underwent WGS with the Ion AmpliSeq SARS-CoV-2 research panel. Among 664 SARS-CoV-2 positive samples, the RT-PCR genotyping assays classified 31.2% as Alpha (N = 207); 48.9% as Delta (N = 325); 19.4% as Omicron (N = 129), 0.3% as Beta (N = 2), and 1 sample as a non-VOC. Matching results were obtained using WGS in 100% of the samples. RT-PCR genotyping assays enable accurate detection of SARS-CoV-2 VOC. Furthermore, they are easily implementable, and the costs and turnaround time are significantly reduced compared to WGS. For this reason, a higher proportion of SARS-CoV-2 positive cases in the VOC surveillance testing can be included, while reserving valuable WGS resources for identification of new variants. Therefore, RT-PCR genotyping assays would be a powerful method to include in SARS-CoV-2 surveillance testing. IMPORTANCE The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) genome changes constantly. It is estimated that there are thousands of variants of SARS-CoV-2 by now. Some of those variants, variants of concern (VOC), pose an increased risk to public health due to higher transmissibility and/or immune escape. Pathogen surveillance helps researchers, epidemiologists, and public health officials to monitor the evolution of infectious diseases agents, alert on the spread of pathogens, and develop counter measures like vaccines. The technique used for the pathogen surveillance is called sequence analysis which makes it possible to examine the building blocks of SARS-CoV-2. In this study, a new PCR method based on the detection of specific changes of those building blocks is presented. This method enables a fast, accurate and cheap determination of different SARS-CoV-2 VOC. Therefore, it would be a powerful method to include in SARS-CoV-2 surveillance testing.
Keywords: RT-PCR genotyping assays; SARS-CoV-2; variant of concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization . 2020. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
-
- 2020. Public Health England. Investigation of SARS-CoV-2 variants of concern. https://www.gov.uk/government/publications/investigation-of-novel-sars-c.... Accessed 29 August 2022.
-
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ, CMMID COVID-19 Working Group . 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science (80-) 372. doi: 10.1126/science.abg3055. - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

