Multiplexed long-read plasmid validation and analysis using OnRamp
- PMID: 37156622
- PMCID: PMC10317119
- DOI: 10.1101/gr.277369.122
Multiplexed long-read plasmid validation and analysis using OnRamp
Abstract
Recombinant plasmid vectors are versatile tools that have facilitated discoveries in molecular biology, genetics, proteomics, and many other fields. As the enzymatic and bacterial processes used to create recombinant DNA can introduce errors, sequence validation is an essential step in plasmid assembly. Sanger sequencing is the current standard for plasmid validation; however, this method is limited by an inability to sequence through complex secondary structure and lacks scalability when applied to full-plasmid sequencing of multiple plasmids owing to read-length limits. Although high-throughput sequencing does provide full-plasmid sequencing at scale, it is impractical and costly when used outside of library-scale validation. Here, we present Oxford nanopore-based rapid analysis of multiplexed plasmids (OnRamp), an alternative method for routine plasmid validation that combines the advantages of high-throughput sequencing's full-plasmid coverage and scalability with Sanger's affordability and accessibility by leveraging nanopore's long-read sequencing technology. We include customized wet-laboratory protocols for plasmid preparation along with a pipeline designed for analysis of read data obtained using these protocols. This analysis pipeline is deployed on the OnRamp web app, which generates alignments between actual and predicted plasmid sequences, quality scores, and read-level views. OnRamp is designed to be broadly accessible regardless of programming experience to facilitate more widespread adoption of long-read sequencing for routine plasmid validation. Here we describe the OnRamp protocols and pipeline and show our ability to obtain full sequences from pooled plasmids while detecting sequence variation even in regions of high secondary structure at less than half the cost of equivalent Sanger sequencing.
© 2023 Mumm et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


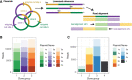
References
-
- Chandak S, Neu J, Tatwawadi K, Mardia J, Lau B, Kubit M, Hulett R, Griffin P, Wootters M, Weissman T, et al. 2020. Overcoming high nanopore basecaller error rates for DNA storage via basecaller-decoder integration and convolutional codes. In ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, pp. 8822–8826. 10.1109/ICASSP40776.2020.9053441 - DOI
-
- Currin A, Swainston N, Dunstan MS, Jervis AJ, Mulherin P, Robinson CJ, Taylor S, Carbonell P, Hollywood KA, Yan C, et al. 2019. Highly multiplexed, fast and accurate nanopore sequencing for verification of synthetic DNA constructs and sequence libraries. Synth Biol 4: ysz025. 10.1093/synbio/ysz025 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
