Single-cell RNA sequencing of liver fine-needle aspirates captures immune diversity in the blood and liver in chronic hepatitis B patients
- PMID: 37158243
- PMCID: PMC10581444
- DOI: 10.1097/HEP.0000000000000438
Single-cell RNA sequencing of liver fine-needle aspirates captures immune diversity in the blood and liver in chronic hepatitis B patients
Abstract
Background and aims: HBV infection is restricted to the liver, where it drives exhaustion of virus-specific T and B cells and pathogenesis through dysregulation of intrahepatic immunity. Our understanding of liver-specific events related to viral control and liver damage has relied almost solely on animal models, and we lack useable peripheral biomarkers to quantify intrahepatic immune activation beyond cytokine measurement. Our objective was to overcome the practical obstacles of liver sampling using fine-needle aspiration and develop an optimized workflow to comprehensively compare the blood and liver compartments within patients with chronic hepatitis B using single-cell RNA sequencing.
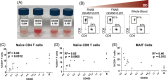
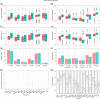
Approach and results: We developed a workflow that enabled multi-site international studies and centralized single-cell RNA sequencing. Blood and liver fine-needle aspirations were collected, and cellular and molecular captures were compared between the Seq-Well S 3 picowell-based and the 10× Chromium reverse-emulsion droplet-based single-cell RNA sequencing technologies. Both technologies captured the cellular diversity of the liver, but Seq-Well S 3 effectively captured neutrophils, which were absent in the 10× dataset. CD8 T cells and neutrophils displayed distinct transcriptional profiles between blood and liver. In addition, liver fine-needle aspirations captured a heterogeneous liver macrophage population. Comparison between untreated patients with chronic hepatitis B and patients treated with nucleoside analogs showed that myeloid cells were highly sensitive to environmental changes while lymphocytes displayed minimal differences.
Conclusions: The ability to electively sample and intensively profile the immune landscape of the liver, and generate high-resolution data, will enable multi-site clinical studies to identify biomarkers for intrahepatic immune activity in HBV and beyond.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
Nádia Conceição-Neto is employed by Janssen Pharmaceutica NV. Jordan Feld consults and received grants from Arbtus, AbbVie, Gilead, GlaxoSmithKline, Janssen, Roche, and Vir. He received grants from Eiger and Enanta. Raymond T. Chung received grants from AbbVie, Boehringer, Bristol Myers Squibb, Gilead, GlaxoSmithKline, Janssen, Merck, and Roche. Robert J. de Knegt consults, is on the speakers' bureau, and received grants from AbbVie, Echosens, and Gilead. He consults and received grants from Bristol Myers Squibb, Janssen, Medtronic, and Roche. He consults for Merck and Norgine. He received grants from GlaxoSmithKline and Phillips. Henry L.A. Janssen consults and received grants from Arbutus, Gilead, GlaxoSmithKline, Janssen, Merck, Roche, and Vir. He consults for Aligos, Antios, Arena, Eiger, Enyo, Regulus, VBI Vaccines, and Viroclinics. He received grants from AbbVie, Arbutus, and Bristol Myers Squibb. Jeroen Aerssens owns stock in Johnson & Johnson. He is employed by Janssen. Jacques Bollekens consults, owns stock in, and holds intellectual property rights with Johnson & Johnson. He is employed by Janssen. Nir Hacohen consults and owns stock in DangerBio. He owns stock in Biontech. Georg M. Lauer consults for and received grants from Janssen. He consults for Altimmune, GlaxoSmithKline, and Roche. Andre Boonstra received grants from Fujirebio, Gilead, GlaxoSmithKline, and Janssen. Alex K. Shalek consults, advises, and owns stock in Cellarity, Dahlia Biosciences, Honeycomb Biotechnologies, Ochre Bio, Repertoire Immune Medicines, Relation Therapeutics, and Santa Ana Bio. He consults and received grants from Janssen. He consults for FL82, Hovione, Merck, Roche, and Third Rock Ventures. He received grants from Leo Pharma A/S and Novo Nordisk. Adam J. Gehring consults and received grants from BlueJay Therapeutics, Gilead, GlaxoSmithKline, Janssen Pharmaceuticals, Roche, and Vir Biotech. He consults for Assembly, Finich Therapeutics, SQ BioTech, Viron Therapeutics, and VBI Vaccines. The remaining authors have no conflicts to report.
Figures





References
-
- Marcellin P, Kutala BK. Liver diseases: a major, neglected global public health problem requiring urgent actions and large-scale screening. Liver Int. 2018;38(Suppl 1):2–6. - PubMed
-
- Polaris Observatory C. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol. 2018;3:383–403. - PubMed
-
- Yuen MF, Chen DS, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, et al. Hepatitis B virus infection. Nat Rev Dis Primers. 2018;4:18035. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

