Crucial role of iron in epigenetic rewriting during adipocyte differentiation mediated by JMJD1A and TET2 activity
- PMID: 37158274
- PMCID: PMC10325906
- DOI: 10.1093/nar/gkad342
Crucial role of iron in epigenetic rewriting during adipocyte differentiation mediated by JMJD1A and TET2 activity
Abstract
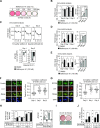
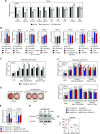
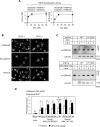
Iron metabolism is closely associated with the pathogenesis of obesity. However, the mechanism of the iron-dependent regulation of adipocyte differentiation remains unclear. Here, we show that iron is essential for rewriting of epigenetic marks during adipocyte differentiation. Iron supply through lysosome-mediated ferritinophagy was found to be crucial during the early stage of adipocyte differentiation, and iron deficiency during this period suppressed subsequent terminal differentiation. This was associated with demethylation of both repressive histone marks and DNA in the genomic regions of adipocyte differentiation-associated genes, including Pparg, which encodes PPARγ, the master regulator of adipocyte differentiation. In addition, we identified several epigenetic demethylases to be responsible for iron-dependent adipocyte differentiation, with the histone demethylase jumonji domain-containing 1A and the DNA demethylase ten-eleven translocation 2 as the major enzymes. The interrelationship between repressive histone marks and DNA methylation was indicated by an integrated genome-wide association analysis, and was also supported by the findings that both histone and DNA demethylation were suppressed by either the inhibition of lysosomal ferritin flux or the knockdown of iron chaperone poly(rC)-binding protein 2. In summary, epigenetic regulations through iron-dependent control of epigenetic enzyme activities play an important role in the organized gene expression mechanisms of adipogenesis.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- de Morais T.R., Gambero A.. Iron chelators in obesity therapy–old drugs from a new perspective?. Eur. J. Pharmacol. 2019; 861:172614. - PubMed
-
- Yun Z., Maecker H.L., Johnson R.S., Giaccia A.J.. Inhibition of PPARγ2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2002; 2:331–341. - PubMed
-
- Lin Q., Lee Y.J., Yun Z.. Differentiation arrest by hypoxia. J. Biol. Chem. 2006; 281:30678–30683. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

